Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment
- PMID: 26170900
- PMCID: PMC4499914
- DOI: 10.1186/s13073-015-0177-8
Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment
Abstract
Background: The human gut microbiome is associated with the development of colon cancer, and recent studies have found changes in the microbiome in cancer patients compared to healthy controls. Studying the microbial communities in the tumor microenvironment may shed light on the role of host-bacteria interactions in colorectal cancer. Here, we highlight the major shifts in the colorectal tumor microbiome relative to that of matched normal colon tissue from the same individual, allowing us to survey the microbial communities in the tumor microenvironment and providing intrinsic control for environmental and host genetic effects on the microbiome.
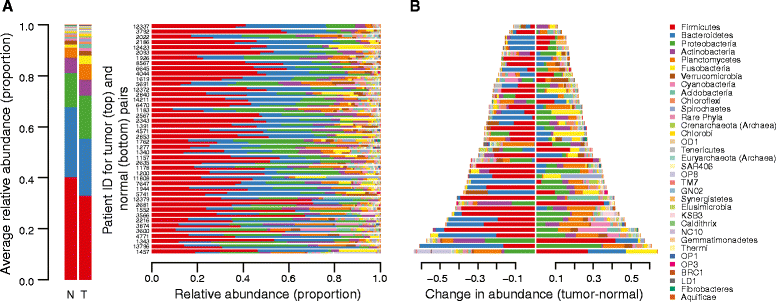
Methods: We sequenced the microbiome in 44 primary tumor and 44 patient-matched normal colon tissue samples to determine differentially abundant microbial taxa These data were also used to functionally characterize the microbiome of the cancer and normal sample pairs and identify functional pathways enriched in the tumor-associated microbiota.
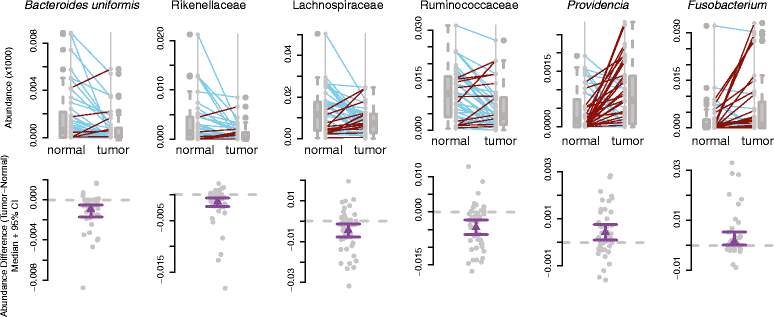
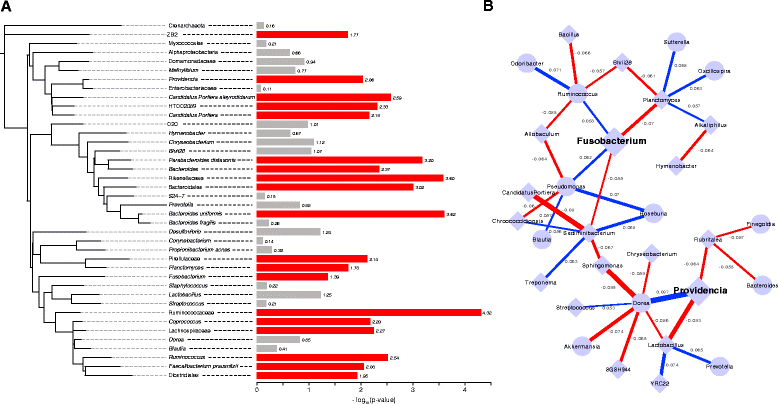
Results: We find that tumors harbor distinct microbial communities compared to nearby healthy tissue. Our results show increased microbial diversity in the tumor microenvironment, with changes in the abundances of commensal and pathogenic bacterial taxa, including Fusobacterium and Providencia. While Fusobacterium has previously been implicated in colorectal cancer, Providencia is a novel tumor-associated agent which has not been identified in previous studies. Additionally, we identified a clear, significant enrichment of predicted virulence-associated genes in the colorectal cancer microenvironment, likely dependent upon the genomes of Fusobacterium and Providencia.
Conclusions: This work identifies bacterial taxa significantly correlated with colorectal cancer, including a novel finding of an elevated abundance of Providencia in the tumor microenvironment. We also describe the predicted metabolic pathways and enzymes differentially present in the tumor-associated microbiome, and show an enrichment of virulence-associated bacterial genes in the tumor microenvironment. This predicted virulence enrichment supports the hypothesis that the microbiome plays an active role in colorectal cancer development and/or progression. Our results provide a starting point for future prognostic and therapeutic research with the potential to improve patient outcomes.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

