Molecular Cloning and Functional Analysis of UV RESISTANCE LOCUS 8 (PeUVR8) from Populus euphratica
- PMID: 26171608
- PMCID: PMC4501546
- DOI: 10.1371/journal.pone.0132390
Molecular Cloning and Functional Analysis of UV RESISTANCE LOCUS 8 (PeUVR8) from Populus euphratica
Abstract
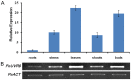
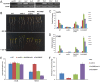
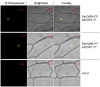
Ultraviolet-B (UV-B; 280-315 nm) light, which is an integral part of the solar radiation reaching the surface of the Earth, induces a broad range of physiological responses in plants. The UV RESISTANCE LOCUS 8 (UVR8) protein is the first and only light photoreceptor characterized to date that is specific for UV-B light and it regulates various aspects of plant growth and development in response to UV-B light. Despite its involvement in the control of important plant traits, most studies on UV-B photoreceptors have focused on Arabidopsis and no data on UVR8 function are available for forest trees. In this study, we isolated a homologue of the UV receptor UVR8 of Arabidopsis, PeUVR8, from Populus euphratica (Euphrates poplar) and analyzed its structure and function in detail. The deduced PeUVR8 amino acid sequence contained nine well-conserved regulator of chromosome condensation 1 (RCC1) repeats and the region 27 amino acids from the C terminus (C27) that interact with COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC1). Secondary and tertiary structure analysis showed that PeUVR8 shares high similarity with the AtUVR8 protein from Arabidopsis thaliana. Using heterologous expression in Arabidopsis, we showed that PeUVR8 overexpression rescued the uvr8 mutant phenotype. In addition, PeUVR8 overexpression in wild-type background seedlings grown under UV-B light inhibited hypocotyl elongation and enhanced anthocyanin accumulation. Furthermore, we examined the interaction between PeUVR8 and AtCOP1 using a bimolecular fluorescence complementation (BiFC) assay. Our data provide evidence that PeUVR8 plays important roles in the control of photomorphogenesis in planta.
Conflict of interest statement
Figures


Similar articles
-
Molecular cloning and functional analysis of a UV-B photoreceptor gene, MdUVR8 (UV Resistance Locus 8), from apple.Plant Sci. 2016 Jun;247:115-26. doi: 10.1016/j.plantsci.2016.03.006. Epub 2016 Mar 17. Plant Sci. 2016. PMID: 27095405
-
Cloning of the cryptochrome-encoding PeCRY1 gene from Populus euphratica and functional analysis in Arabidopsis.PLoS One. 2014 Dec 12;9(12):e115201. doi: 10.1371/journal.pone.0115201. eCollection 2014. PLoS One. 2014. PMID: 25503486 Free PMC article.
-
Expression of Tomato UVR8 in Arabidopsis reveals conserved photoreceptor function.Plant Sci. 2021 Feb;303:110766. doi: 10.1016/j.plantsci.2020.110766. Epub 2020 Nov 21. Plant Sci. 2021. PMID: 33487351
-
The UV-B photoreceptor UVR8: from structure to physiology.Plant Cell. 2014 Jan;26(1):21-37. doi: 10.1105/tpc.113.119446. Epub 2014 Jan 30. Plant Cell. 2014. PMID: 24481075 Free PMC article. Review.
-
Perception and Signaling of Ultraviolet-B Radiation in Plants.Annu Rev Plant Biol. 2021 Jun 17;72:793-822. doi: 10.1146/annurev-arplant-050718-095946. Epub 2021 Feb 26. Annu Rev Plant Biol. 2021. PMID: 33636992 Review.
Cited by
-
A seed-specific transcription factor, HSFA9, anticipates UV-B light responses by mimicking the activation of the UV-B receptor in tobacco.Plant J. 2022 Sep;111(5):1439-1452. doi: 10.1111/tpj.15901. Epub 2022 Aug 1. Plant J. 2022. PMID: 35811570 Free PMC article.
-
Tomato UV-B receptor SlUVR8 mediates plant acclimation to UV-B radiation and enhances fruit chloroplast development via regulating SlGLK2.Sci Rep. 2018 Apr 17;8(1):6097. doi: 10.1038/s41598-018-24309-y. Sci Rep. 2018. PMID: 29666396 Free PMC article.
-
Comparative transcription analysis of photosensitive and non-photosensitive eggplants to identify genes involved in dark regulated anthocyanin synthesis.BMC Genomics. 2019 Aug 28;20(1):678. doi: 10.1186/s12864-019-6023-4. BMC Genomics. 2019. PMID: 31455222 Free PMC article.
-
On the natural diversity of phenylacylated-flavonoid and their in planta function under conditions of stress.Phytochem Rev. 2018;17(2):279-290. doi: 10.1007/s11101-017-9531-3. Epub 2017 Sep 8. Phytochem Rev. 2018. PMID: 29755304 Free PMC article. Review.
-
Hydrogen peroxide, nitric oxide and UV RESISTANCE LOCUS8 interact to mediate UV-B-induced anthocyanin biosynthesis in radish sprouts.Sci Rep. 2016 Jul 12;6:29164. doi: 10.1038/srep29164. Sci Rep. 2016. PMID: 27404993 Free PMC article.
References
-
- Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Developmental Biology 260: 289–297. - PubMed
-
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D (1995) Phytochromes: photosensory perception and signal transduction. Science 268: 675–680. - PubMed
-
- Briggs WR, Huala E (1999) Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol 15: 33–62. - PubMed
-
- Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: Blue light receptors for plants and animals. Science 284: 760–765. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

