Near universal detection of alterations in CTNNB1 and Wnt pathway regulators in desmoid-type fibromatosis by whole-exome sequencing and genomic analysis
- PMID: 26171757
- PMCID: PMC4548882
- DOI: 10.1002/gcc.22272
Near universal detection of alterations in CTNNB1 and Wnt pathway regulators in desmoid-type fibromatosis by whole-exome sequencing and genomic analysis
Abstract
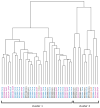
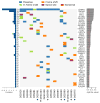
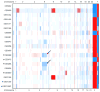
CTNNB1 mutations or APC abnormalities have been observed in ∼85% of desmoids examined by Sanger sequencing and are associated with Wnt/β-catenin activation. We sought to identify molecular aberrations in "wild-type" tumors (those without CTNNB1 or APC alteration) and to determine their prognostic relevance. CTNNB1 was examined by Sanger sequencing in 117 desmoids; a mutation was observed in 101 (86%) and 16 were wild type. Wild-type status did not associate with tumor recurrence. Moreover, in unsupervised clustering based on U133A-derived gene expression profiles, wild-type and mutated tumors clustered together. Whole-exome sequencing of eight of the wild-type desmoids revealed that three had a CTNNB1 mutation that had been undetected by Sanger sequencing. The mutation was found in a mean 16% of reads (vs. 37% for mutations identified by Sanger). Of the other five wild-type tumors sequenced, two had APC loss, two had chromosome 6 loss, and one had mutation of BMI1. The finding of low-frequency CTNNB1 mutation or APC loss in wild-type desmoids was validated in the remaining eight wild-type desmoids; directed miSeq identified low-frequency CTNNB1 mutation in four and comparative genomic hybridization identified APC loss in one. These results demonstrate that mutations affecting CTNNB1 or APC occur more frequently in desmoids than previously recognized (111 of 117; 95%), and designation of wild-type genotype is largely determined by sensitivity of detection methods. Even true CTNNB1 wild-type tumors (determined by next-generation sequencing) may have genomic alterations associated with Wnt activation (chromosome 6 loss/BMI1 mutation), supporting Wnt/β-catenin activation as the common pathway governing desmoid initiation.
© 2015 Wiley Periodicals, Inc.
Figures
References
-
- Alman BA, Pajerski ME, Diaz-Cano S, Corboy K, Wolfe HJ. Aggressive fibromatosis (desmoid tumor) is a monoclonal disorder. Diagn Mol Pathol. 1997a;6:98–101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

