Multifaceted interactions and regulation between antizyme and its interacting proteins cyclin D1, ornithine decarboxylase and antizyme inhibitor
- PMID: 26172301
- PMCID: PMC4695161
- DOI: 10.18632/oncotarget.4469
Multifaceted interactions and regulation between antizyme and its interacting proteins cyclin D1, ornithine decarboxylase and antizyme inhibitor
Abstract
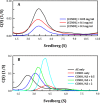
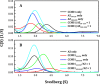
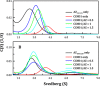
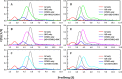
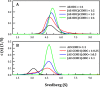
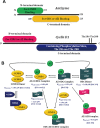
Ornithine decarboxylase (ODC), cyclin D1 (CCND1) and antizyme inhibitor (AZI) promote cell growth. ODC and CCND1 can be degraded through antizyme (AZ)-mediated 26S proteasomal degradation. This paper describes a mechanistic study of the molecular interactions between AZ and its interacting proteins. The dissociation constant (Kd) of the binary AZ-CCND1 complex and the respective binding sites of AZ and CCND1 were determined. Our data indicate that CCND1 has a 4-fold lower binding affinity for AZ than does ODC and an approximately 40-fold lower binding affinity for AZ than does AZI. The Kd values of AZ-CCND1, AZ-ODC and AZ-AZI were 0.81, 0.21 and 0.02 μM, respectively. Furthermore, the Kd values for CCND1 binding to the AZ N-terminal peptide (AZ34-124) and AZ C-terminal peptide (AZ100-228) were 0.92 and 8.97 μM, respectively, indicating that the binding site of CCND1 may reside at the N-terminus of AZ, rather than the C-terminus. Our data also show that the ODC-AZ-CCND1 ternary complex may exist in equilibrium. The Kd values of the [AZ-CCND1]-ODC and [AZ-ODC]-CCND1 complexes were 1.26 and 4.93 μM, respectively. This is the first paper to report the reciprocal regulation of CCND1 and ODC through AZ-dependent 26S proteasomal degradation.
Keywords: biochemistry; cell cycle; molecular and cellular biology; oncogene; signal transduction.
Figures

Similar articles
-
Degradation of antizyme inhibitor, an ornithine decarboxylase homologous protein, is ubiquitin-dependent and is inhibited by antizyme.J Biol Chem. 2004 Dec 24;279(52):54097-102. doi: 10.1074/jbc.M410234200. Epub 2004 Oct 18. J Biol Chem. 2004. PMID: 15491992
-
Critical factors governing the difference in antizyme-binding affinities between human ornithine decarboxylase and antizyme inhibitor.PLoS One. 2011 Apr 28;6(4):e19253. doi: 10.1371/journal.pone.0019253. PLoS One. 2011. PMID: 21552531 Free PMC article.
-
Minimal antizyme peptide fully functioning in the binding and inhibition of ornithine decarboxylase and antizyme inhibitor.PLoS One. 2011;6(9):e24366. doi: 10.1371/journal.pone.0024366. Epub 2011 Sep 9. PLoS One. 2011. PMID: 21931692 Free PMC article.
-
Degradation of ornithine decarboxylase by the 26S proteasome.Biochem Biophys Res Commun. 2000 Jan 7;267(1):1-6. doi: 10.1006/bbrc.1999.1706. Biochem Biophys Res Commun. 2000. PMID: 10623564 Review.
-
Regulation of cellular polyamine levels and cellular proliferation by antizyme and antizyme inhibitor.Essays Biochem. 2009 Nov 4;46:47-61. doi: 10.1042/bse0460004. Essays Biochem. 2009. PMID: 20095969 Review.
Cited by
-
Elevated Polyamines in Saliva of Pancreatic Cancer.Cancers (Basel). 2018 Feb 5;10(2):43. doi: 10.3390/cancers10020043. Cancers (Basel). 2018. PMID: 29401744 Free PMC article.
-
Role of Antizyme Inhibitor Proteins in Cancers and Beyond.Onco Targets Ther. 2021 Jan 25;14:667-682. doi: 10.2147/OTT.S281157. eCollection 2021. Onco Targets Ther. 2021. PMID: 33531815 Free PMC article. Review.
-
Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use.Nucleic Acids Res. 2016 Sep 6;44(15):7007-78. doi: 10.1093/nar/gkw530. Epub 2016 Jul 19. Nucleic Acids Res. 2016. PMID: 27436286 Free PMC article. Review.
-
C-Methylated Spermidine Derivatives: Convenient Syntheses and Antizyme-Related Effects.Biomolecules. 2023 May 31;13(6):916. doi: 10.3390/biom13060916. Biomolecules. 2023. PMID: 37371496 Free PMC article.
-
Antizyme inhibitor 1: a potential carcinogenic molecule.Cancer Sci. 2017 Feb;108(2):163-169. doi: 10.1111/cas.13122. Cancer Sci. 2017. PMID: 27870265 Free PMC article. Review.
References
-
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. - PubMed
-
- Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. - PubMed
-
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

