The Contribution of Missense Mutations in Core and Rim Residues of Protein-Protein Interfaces to Human Disease
- PMID: 26173036
- PMCID: PMC4548493
- DOI: 10.1016/j.jmb.2015.07.004
The Contribution of Missense Mutations in Core and Rim Residues of Protein-Protein Interfaces to Human Disease
Abstract
Missense mutations at protein-protein interaction sites, called interfaces, are important contributors to human disease. Interfaces are non-uniform surface areas characterized by two main regions, "core" and "rim", which differ in terms of evolutionary conservation and physicochemical properties. Moreover, within interfaces, only a small subset of residues ("hot spots") is crucial for the binding free energy of the protein-protein complex. We performed a large-scale structural analysis of human single amino acid variations (SAVs) and demonstrated that disease-causing mutations are preferentially located within the interface core, as opposed to the rim (p<0.01). In contrast, the interface rim is significantly enriched in polymorphisms, similar to the remaining non-interacting surface. Energetic hot spots tend to be enriched in disease-causing mutations compared to non-hot spots (p=0.05), regardless of their occurrence in core or rim residues. For individual amino acids, the frequency of substitution into a polymorphism or disease-causing mutation differed to other amino acids and was related to its structural location, as was the type of physicochemical change introduced by the SAV. In conclusion, this study demonstrated the different distribution and properties of disease-causing SAVs and polymorphisms within different structural regions and in relation to the energetic contribution of amino acid in protein-protein interfaces, thus highlighting the importance of a structural system biology approach for predicting the effect of SAVs.
Keywords: SAVs; core and rim interface; human disease; nsSNPs; protein–protein interaction.
Copyright © 2015. Published by Elsevier Ltd.
Figures


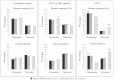

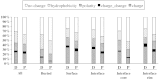
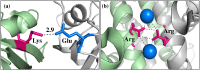
Similar articles
-
Analysis of single amino acid variations in singlet hot spots of protein-protein interfaces.Bioinformatics. 2018 Sep 1;34(17):i795-i801. doi: 10.1093/bioinformatics/bty569. Bioinformatics. 2018. PMID: 30423104
-
Protein-protein interaction sites are hot spots for disease-associated nonsynonymous SNPs.Hum Mutat. 2012 Feb;33(2):359-63. doi: 10.1002/humu.21656. Epub 2011 Dec 27. Hum Mutat. 2012. PMID: 22072597
-
Structural and Computational Characterization of Disease-Related Mutations Involved in Protein-Protein Interfaces.Int J Mol Sci. 2019 Mar 29;20(7):1583. doi: 10.3390/ijms20071583. Int J Mol Sci. 2019. PMID: 30934865 Free PMC article.
-
Approaches and resources for prediction of the effects of non-synonymous single nucleotide polymorphism on protein function and interactions.Curr Pharm Biotechnol. 2008 Apr;9(2):123-33. doi: 10.2174/138920108783955164. Curr Pharm Biotechnol. 2008. PMID: 18393868 Review.
-
Anatomy of hot spots in protein interfaces.J Mol Biol. 1998 Jul 3;280(1):1-9. doi: 10.1006/jmbi.1998.1843. J Mol Biol. 1998. PMID: 9653027 Review.
Cited by
-
Comprehensive characterization of amino acid positions in protein structures reveals molecular effect of missense variants.Proc Natl Acad Sci U S A. 2020 Nov 10;117(45):28201-28211. doi: 10.1073/pnas.2002660117. Epub 2020 Oct 26. Proc Natl Acad Sci U S A. 2020. PMID: 33106425 Free PMC article.
-
Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated?J Mol Biol. 2019 May 17;431(11):2197-2212. doi: 10.1016/j.jmb.2019.04.009. Epub 2019 Apr 14. J Mol Biol. 2019. PMID: 30995449 Free PMC article.
-
Co-evolutionary landscape at the interface and non-interface regions of protein-protein interaction complexes.Comput Struct Biotechnol J. 2021 Jun 24;19:3779-3795. doi: 10.1016/j.csbj.2021.06.039. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34285778 Free PMC article.
-
Mutational survivorship bias: The case of PNKP.PLoS One. 2020 Dec 17;15(12):e0237682. doi: 10.1371/journal.pone.0237682. eCollection 2020. PLoS One. 2020. PMID: 33332469 Free PMC article.
-
PROT-ON: A structure-based detection of designer PROTein interface MutatiONs.Front Mol Biosci. 2023 Mar 1;10:1063971. doi: 10.3389/fmolb.2023.1063971. eCollection 2023. Front Mol Biosci. 2023. PMID: 36936988 Free PMC article.
References
-
- David A., Razali R., Wass M.N., Sternberg M.J.E. Protein–protein interaction sites are hot spots for disease-associated nonsynonymous SNPs. Hum. Mutat. 2012;33:359–363. - PubMed
-
- Alexov E., Sternberg M. Understanding molecular effects of naturally occurring genetic differences. J. Mol. Biol. 2013;425:3911–3913. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- dbSNP/RS201053197
- dbSNP/RS34116584
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

