Emergence of a New Highly Successful Acapsular Group A Streptococcus Clade of Genotype emm89 in the United Kingdom
- PMID: 26173696
- PMCID: PMC4502227
- DOI: 10.1128/mBio.00622-15
Emergence of a New Highly Successful Acapsular Group A Streptococcus Clade of Genotype emm89 in the United Kingdom
Abstract
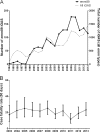
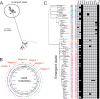
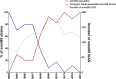
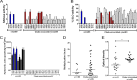
Group A Streptococcus (GAS) genotype emm89 is increasingly recognized as a leading cause of disease worldwide, yet factors that underlie the success of this emm type are unknown. Surveillance identified a sustained nationwide increase in emm89 invasive GAS disease in the United Kingdom, prompting longitudinal investigation of this genotype. Whole-genome sequencing revealed a recent dramatic shift in the emm89 population with the emergence of a new clade that increased to dominance over previous emm89 variants. Temporal analysis indicated that the clade arose in the early 1990s but abruptly increased in prevalence in 2008, coinciding with an increased incidence of emm89 infections. Although standard variable typing regions (emm subtype, tee type, sof type, and multilocus sequence typing [MLST]) remained unchanged, uniquely the emergent clade had undergone six distinct regions of homologous recombination across the genome compared to the rest of the sequenced emm89 population. Two of these regions affected known virulence factors, the hyaluronic acid capsule and the toxins NADase and streptolysin O. Unexpectedly, and in contrast to the rest of the sequenced emm89 population, the emergent clade-associated strains were genetically acapsular, rendering them unable to produce the hyaluronic acid capsule. The emergent clade-associated strains had also acquired an NADase/streptolysin O locus nearly identical to that found in emm12 and modern emm1 strains but different from the rest of the sequenced emm89 population. The emergent clade-associated strains had enhanced expression of NADase and streptolysin O. The genome remodeling in the new clade variant and the resultant altered phenotype appear to have conferred a selective advantage over other emm89 variants and may explain the changes observed in emm89 GAS epidemiology.
Importance: Sudden upsurges or epidemic waves are common features of group A streptococcal disease. Although the mechanisms behind such changes are largely unknown, they are often associated with an expansion of a single genotype within the population. Using whole-genome sequencing, we investigated a nationwide increase in invasive disease caused by the genotype emm89 in the United Kingdom. We identified a new clade variant that had recently emerged in the emm89 population after having undergone several core genomic recombination-related changes, two of which affected known virulence factors. An unusual finding of the new variant was the loss of the hyaluronic acid capsule, previously thought to be essential for causing invasive disease. A further genomic adaptation in the NADase/streptolysin O locus resulted in enhanced production of these toxins. Recombination-related genome remodeling is clearly an important mechanism in group A Streptococcus that can give rise to more successful and potentially more pathogenic variants.
Copyright © 2015 Turner et al.
Figures

Comment in
-
Emergence of the Same Successful Clade among Distinct Populations of emm89 Streptococcus pyogenes in Multiple Geographic Regions.mBio. 2015 Dec 1;6(6):e01780-15. doi: 10.1128/mBio.01780-15. mBio. 2015. PMID: 26628724 Free PMC article. No abstract available.
-
Turner et al. Reply to "Emergence of the Same Successful Clade among Distinct Populations of emm89 Streptococcus pyogenes in Multiple Geographic Regions".mBio. 2015 Dec 1;6(6):e01883-15. doi: 10.1128/mBio.01883-15. mBio. 2015. PMID: 26628727 Free PMC article. No abstract available.
Similar articles
-
The Emergence of Successful Streptococcus pyogenes Lineages through Convergent Pathways of Capsule Loss and Recombination Directing High Toxin Expression.mBio. 2019 Dec 10;10(6):e02521-19. doi: 10.1128/mBio.02521-19. mBio. 2019. PMID: 31822586 Free PMC article.
-
Trading Capsule for Increased Cytotoxin Production: Contribution to Virulence of a Newly Emerged Clade of emm89 Streptococcus pyogenes.mBio. 2015 Oct 6;6(5):e01378-15. doi: 10.1128/mBio.01378-15. mBio. 2015. PMID: 26443457 Free PMC article.
-
Population and Whole Genome Sequence Based Characterization of Invasive Group A Streptococci Recovered in the United States during 2015.mBio. 2017 Sep 19;8(5):e01422-17. doi: 10.1128/mBio.01422-17. mBio. 2017. PMID: 28928212 Free PMC article.
-
Tissue tropisms in group A streptococcal infections.Future Microbiol. 2010 Apr;5(4):623-38. doi: 10.2217/fmb.10.28. Future Microbiol. 2010. PMID: 20353302 Free PMC article. Review.
-
New understandings in Streptococcus pyogenes.Curr Opin Infect Dis. 2011 Jun;24(3):196-202. doi: 10.1097/QCO.0b013e3283458f7e. Curr Opin Infect Dis. 2011. PMID: 21415743 Review.
Cited by
-
Molecular characterization of Streptococcus pyogenes (StrepA) non-invasive isolates during the 2022-2023 UK upsurge.Microb Genom. 2024 Aug;10(8):001277. doi: 10.1099/mgen.0.001277. Microb Genom. 2024. PMID: 39133528 Free PMC article.
-
Epidemiology of invasive group A streptococcal infections in Norway 2010-2014: A retrospective cohort study.Eur J Clin Microbiol Infect Dis. 2016 Oct;35(10):1639-48. doi: 10.1007/s10096-016-2704-y. Epub 2016 Jun 16. Eur J Clin Microbiol Infect Dis. 2016. PMID: 27311458
-
Canada-Wide Epidemic of emm74 Group A Streptococcus Invasive Disease.Open Forum Infect Dis. 2018 Apr 20;5(5):ofy085. doi: 10.1093/ofid/ofy085. eCollection 2018 May. Open Forum Infect Dis. 2018. PMID: 29780850 Free PMC article.
-
Reduced In Vitro Susceptibility of Streptococcus pyogenes to β-Lactam Antibiotics Associated with Mutations in the pbp2x Gene Is Geographically Widespread.J Clin Microbiol. 2020 Mar 25;58(4):e01993-19. doi: 10.1128/JCM.01993-19. Print 2020 Mar 25. J Clin Microbiol. 2020. PMID: 31996443 Free PMC article.
-
Transcriptome Remodeling Contributes to Epidemic Disease Caused by the Human Pathogen Streptococcus pyogenes.mBio. 2016 May 31;7(3):e00403-16. doi: 10.1128/mBio.00403-16. mBio. 2016. PMID: 27247229 Free PMC article.
References
-
- Shea PR, Ewbank AL, Gonzalez-Lugo JH, Martagon-Rosado AJ, Martinez-Gutierrez JC, Rehman HA, Serrano-Gonzalez M, Fittipaldi N, Beres SB, Flores AR, Low DE, Willey BM, Musser JM. 2011. Group A Streptococcus emm gene types in pharyngeal isolates, Ontario, Canada, 2002–2010. Emerg Infect Dis 17:2010–2017. doi:10.3201/eid1711.110159. - DOI - PMC - PubMed
-
- Friães A, Pinto FR, Silva-Costa C, Ramirez M, Melo-Cristino J, The Portuguese Group for the Study of Streptococcal Infections . 2012. Group A streptococci clones associated with invasive infections and pharyngitis in Portugal present differences in emm types, superantigen gene content and antimicrobial resistance. BMC Microbiol 12:280. doi:10.1186/1471-2180-12-280. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

