Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile
- PMID: 26173701
- PMCID: PMC4502226
- DOI: 10.1128/mBio.00974-15
Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile
Abstract
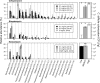
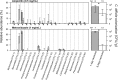
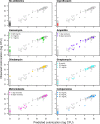
Perturbations to the gut microbiota can result in a loss of colonization resistance against gastrointestinal pathogens such as Clostridium difficile. Although C. difficile infection is commonly associated with antibiotic use, the precise alterations to the microbiota associated with this loss in function are unknown. We used a variety of antibiotic perturbations to generate a diverse array of gut microbiota structures, which were then challenged with C. difficile spores. Across these treatments we observed that C. difficile resistance was never attributable to a single organism, but rather it was the result of multiple microbiota members interacting in a context-dependent manner. Using relative abundance data, we built a machine learning regression model to predict the levels of C. difficile that were found 24 h after challenging the perturbed communities. This model was able to explain 77.2% of the variation in the observed number of C. difficile per gram of feces. This model revealed important bacterial populations within the microbiota, which correlation analysis alone did not detect. Specifically, we observed that populations associated with the Porphyromonadaceae, Lachnospiraceae, Lactobacillus, and Alistipes were protective and populations associated with Escherichia and Streptococcus were associated with high levels of colonization. In addition, a population affiliated with the Akkermansia indicated a strong context dependency on other members of the microbiota. Together, these results indicate that individual bacterial populations do not drive colonization resistance to C. difficile. Rather, multiple diverse assemblages act in concert to mediate colonization resistance.
Importance: The gastrointestinal tract harbors a complex community of bacteria, known as the microbiota, which plays an integral role preventing its colonization by gut pathogens. This resistance has been shown to be crucial for protection against Clostridium difficile infections (CDI), which are the leading source of hospital-acquired infections in the United States. Antibiotics are a major risk factor for acquiring CDI due to their effect on the normal structure of the indigenous gut microbiota. We found that diverse antibiotic perturbations gave rise to altered communities that varied in their susceptibility to C. difficile colonization. We found that multiple coexisting populations, not one specific population of bacteria, conferred resistance. By understanding the relationships between C. difficile and members of the microbiota, it will be possible to better manage this important infection.
Copyright © 2015 Schubert et al.
Figures



Comment in
-
Modeling the Context-Dependent Associations between the Gut Microbiome, Its Environment, and Host Health.mBio. 2015 Sep 8;6(5):e01367-15. doi: 10.1128/mBio.01367-15. mBio. 2015. PMID: 26350971 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
