DNA Methylation in Basal Metazoans: Insights from Ctenophores
- PMID: 26173712
- PMCID: PMC4817592
- DOI: 10.1093/icb/icv086
DNA Methylation in Basal Metazoans: Insights from Ctenophores
Abstract
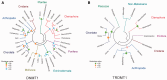
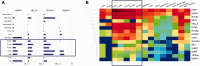
Epigenetic modifications control gene expression without altering the primary DNA sequence. However, little is known about DNA methylation in invertebrates and its evolution. Here, we characterize two types of genomic DNA methylation in ctenophores, 5-methyl cytosine (5-mC) and the unconventional form of methylation 6-methyl adenine (6-mA). Using both bisulfite sequencing and an ELISA-based colorimetric assay, we experimentally confirmed the presence of 5-mC DNA methylation in ctenophores. In contrast to other invertebrates studied, Mnemiopsis leidyi has lower levels of genome-wide 5-mC methylation, but higher levels of 5-mC methylation in promoters when compared with gene bodies. Phylogenetic analysis showed that ctenophores have distinct forms of DNA methyltransferase 1 (DNMT1); the zf-CXXC domain type, which localized DNMT1 to CpG sites, and is a metazoan specific innovation. We also show that ctenophores encode the full repertoire of putative enzymes for 6-mA DNA methylation, and these genes are expressed in the aboral organ of Mnemiopsis. Using an ELISA-based colorimetric assay, we experimentally confirmed the presence of 6-mA methylation in the genomes of three different species of ctenophores, M. leidyi, Beroe abyssicola, and Pleurobrachia bachei. The functional role of this novel epigenomic mark is currently unknown. In summary, despite their compact genomes, there is a wide variety of epigenomic mechanisms employed by basal metazoans that provide novel insights into the evolutionary origins of biological novelties.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Integrative and Comparative Biology. All rights reserved. For permissions please email: journals.permissions@oup.com.
Figures




Similar articles
-
Stage-specific DNA methylation dynamics in mammalian heart development.Epigenomics. 2025 Apr;17(5):359-371. doi: 10.1080/17501911.2025.2467024. Epub 2025 Feb 21. Epigenomics. 2025. PMID: 39980349 Review.
-
[Epigenetics' implication in autism spectrum disorders: A review].Encephale. 2017 Aug;43(4):374-381. doi: 10.1016/j.encep.2016.07.007. Epub 2016 Sep 28. Encephale. 2017. PMID: 27692350 French.
-
CGGBP1 from higher amniotes restricts cytosine methylation and drives a GC-bias in transcription factor-binding sites at repressed promoters.Transcription. 2025 Jul 31:1-36. doi: 10.1080/21541264.2025.2533598. Online ahead of print. Transcription. 2025. PMID: 40740140
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
SNP-associated differential methylation in ARHGEF38: insights into genetic-epigenetic interactions.Epigenomics. 2025 Jun;17(9):579-588. doi: 10.1080/17501911.2025.2513215. Epub 2025 May 30. Epigenomics. 2025. PMID: 40444651
Cited by
-
Detection of N6-Methyladenine in Eukaryotes.Adv Exp Med Biol. 2021;1280:83-95. doi: 10.1007/978-3-030-51652-9_6. Adv Exp Med Biol. 2021. PMID: 33791976
-
A Broad Survey of Gene Body and Repeat Methylation in Cnidaria Reveals a Complex Evolutionary History.Genome Biol Evol. 2022 Feb 4;14(2):evab284. doi: 10.1093/gbe/evab284. Genome Biol Evol. 2022. PMID: 35104341 Free PMC article.
-
Genomic data do not support comb jellies as the sister group to all other animals.Proc Natl Acad Sci U S A. 2015 Dec 15;112(50):15402-7. doi: 10.1073/pnas.1518127112. Epub 2015 Nov 30. Proc Natl Acad Sci U S A. 2015. PMID: 26621703 Free PMC article.
-
Effects of a parental exposure to diuron on Pacific oyster spat methylome.Environ Epigenet. 2017 Apr 19;3(1):dvx004. doi: 10.1093/eep/dvx004. eCollection 2017 Jan. Environ Epigenet. 2017. PMID: 29492306 Free PMC article.
-
DNA Methylation in Ctenophores.Methods Mol Biol. 2024;2757:447-460. doi: 10.1007/978-1-0716-3642-8_18. Methods Mol Biol. 2024. PMID: 38668978
References
-
- Ahmad M, Tuteja R. 2013. Plasmodium falciparum RuvB1 is an active DNA helicase and translocates in the 5′–3′ direction. Gene 515:99–109. - PubMed
-
- Albalat R. 2008. Evolution of DNA-methylation machinery: DNA methyltransferases and methyl-DNA binding proteins in the amphioxus Branchiostoma floridae. Dev Genes Evol 218:691–701. - PubMed
-
- Bigey P, Ramchandani S, Theberge J, Araujo FD, Szyf M. 2000. Transcriptional regulation of the human DNA methyltransferase (dnmt1) gene. Gene 242:407–18. - PubMed
-
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16:6–21. - PubMed
-
- Bird A. 2007. Perceptions of epigenetics. Nature 447:396–8. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

