De novo assembly and characterization of central nervous system transcriptome reveals neurotransmitter signaling systems in the rice striped stem borer, Chilo suppressalis
- PMID: 26173787
- PMCID: PMC4501067
- DOI: 10.1186/s12864-015-1742-7
De novo assembly and characterization of central nervous system transcriptome reveals neurotransmitter signaling systems in the rice striped stem borer, Chilo suppressalis
Abstract
Background: Neurotransmitter signaling systems play crucial roles in multiple physiological and behavioral processes in insects. Genome wide analyses of de novo transcriptome sequencing and gene specific expression profiling provide rich resources for studying neurotransmitter signaling pathways. The rice striped stem borer, Chilo suppressalis is a destructive rice pest in China and other Asian countries. The characterization of genes involved in neurotransmitter biosynthesis and transport could identify potential targets for disruption of the neurochemical communication and for crop protection.
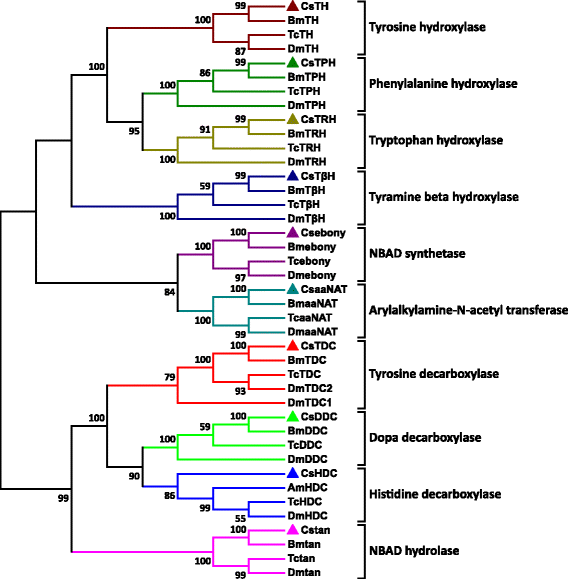
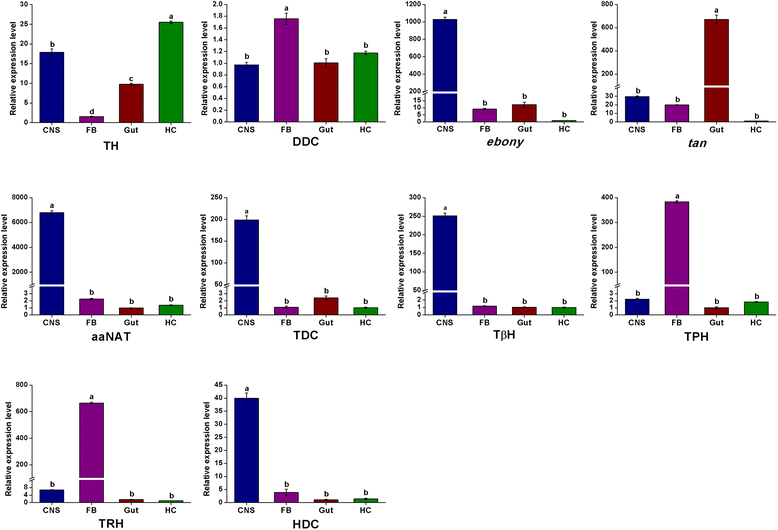
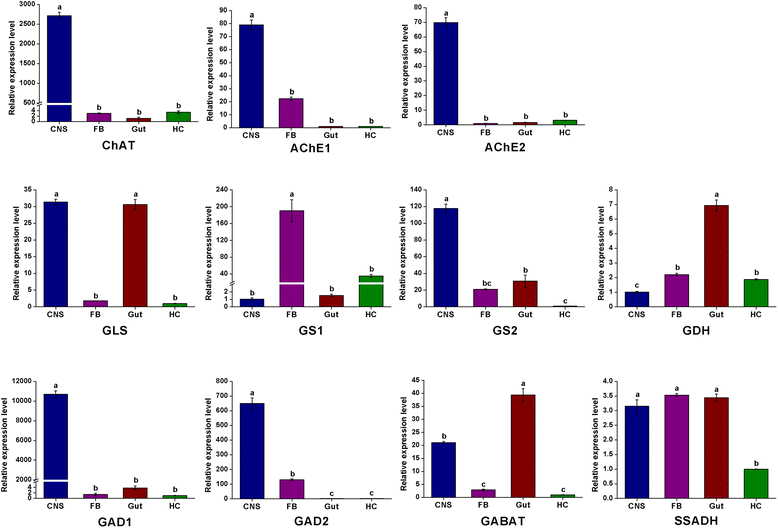
Results: Here we report de novo sequencing of the C. suppressalis central nervous system transcriptome, identification and expression profiles of genes putatively involved in neurotransmitter biosynthesis, packaging, and recycling/degradation. A total of 54,411 unigenes were obtained from the transcriptome analysis. Among these unigenes, we have identified 32 unigenes (31 are full length genes), which encode 21 enzymes and 11 transporters putatively associated with biogenic aminergic signaling, acetylcholinergic signaling, glutamatergic signaling and GABAergic signaling. RT-PCR and qRT-PCR results indicated that 12 enzymes were highly expressed in the central nervous system and all the transporters were expressed at significantly high levels in the central nervous system. In addition, the transcript abundances of enzymes and transporters in the central nervous system were validated by qRT-PCR. The high expression levels of these genes suggest their important roles in the central nervous system.
Conclusions: Our study identified genes potentially involved in neurotransmitter biosynthesis and transport in C. suppressalis and these genes could serve as targets to interfere with neurotransmitter production. This study presents an opportunity for the development of specific and environmentally safe insecticides for pest control.
Figures











Similar articles
-
Identification and expression profiles of neuropeptides and their G protein-coupled receptors in the rice stem borer Chilo suppressalis.Sci Rep. 2016 Jun 29;6:28976. doi: 10.1038/srep28976. Sci Rep. 2016. PMID: 27353701 Free PMC article.
-
Genome-wide analysis reveals the expansion of Cytochrome P450 genes associated with xenobiotic metabolism in rice striped stem borer, Chilo suppressalis.Biochem Biophys Res Commun. 2014 Jan 10;443(2):756-60. doi: 10.1016/j.bbrc.2013.12.045. Epub 2013 Dec 17. Biochem Biophys Res Commun. 2014. PMID: 24361403
-
Combined transcriptome and metabolome analyses to understand the dynamic responses of rice plants to attack by the rice stem borer Chilo suppressalis (Lepidoptera: Crambidae).BMC Plant Biol. 2016 Dec 7;16(1):259. doi: 10.1186/s12870-016-0946-6. BMC Plant Biol. 2016. PMID: 27923345 Free PMC article.
-
Jasmonic Acid (JA) Signaling Pathway in Rice Defense Against Chilo suppressalis Infestation.Rice (N Y). 2025 Feb 18;18(1):7. doi: 10.1186/s12284-025-00761-z. Rice (N Y). 2025. PMID: 39964588 Free PMC article. Review.
-
Defense Strategies of Rice in Response to the Attack of the Herbivorous Insect, Chilo suppressalis.Int J Mol Sci. 2023 Sep 21;24(18):14361. doi: 10.3390/ijms241814361. Int J Mol Sci. 2023. PMID: 37762665 Free PMC article. Review.
Cited by
-
Molecular and Pharmacological Characterization of β-Adrenergic-like Octopamine Receptors in the Endoparasitoid Cotesia chilonis (Hymenoptera: Braconidae).Int J Mol Sci. 2022 Nov 22;23(23):14513. doi: 10.3390/ijms232314513. Int J Mol Sci. 2022. PMID: 36498840 Free PMC article.
-
Identification and expression profiles of neuropeptides and their G protein-coupled receptors in the rice stem borer Chilo suppressalis.Sci Rep. 2016 Jun 29;6:28976. doi: 10.1038/srep28976. Sci Rep. 2016. PMID: 27353701 Free PMC article.
-
Characterization of the Adult Head Transcriptome and Identification of Migration and Olfaction Genes in the Oriental Armyworm Mythimna separate.Sci Rep. 2017 May 24;7(1):2324. doi: 10.1038/s41598-017-02513-6. Sci Rep. 2017. PMID: 28539591 Free PMC article.
-
A Possible Role of Crustacean Cardioactive Peptide in Regulating Immune Response in Hepatopancreas of Mud Crab.Front Immunol. 2020 Apr 30;11:711. doi: 10.3389/fimmu.2020.00711. eCollection 2020. Front Immunol. 2020. PMID: 32425935 Free PMC article.
-
ACE: an efficient and sensitive tool to detect insecticide resistance-associated mutations in insect acetylcholinesterase from RNA-Seq data.BMC Bioinformatics. 2017 Jul 10;18(1):330. doi: 10.1186/s12859-017-1741-6. BMC Bioinformatics. 2017. PMID: 28693417 Free PMC article.
References
-
- McCoole MD, D'Andrea BT, Baer KN, Christie AE. Genomic analyses of gas (nitric oxide and carbon monoxide) and small molecule transmitter (acetylcholine, glutamate and GABA) signaling systems in Daphnia pulex. Comp Biochem Phys D. 2012;7:124–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

