Genes conserved in bilaterians but jointly lost with Myc during nematode evolution are enriched in cell proliferation and cell migration functions
- PMID: 26173873
- PMCID: PMC4568025
- DOI: 10.1007/s00427-015-0508-1
Genes conserved in bilaterians but jointly lost with Myc during nematode evolution are enriched in cell proliferation and cell migration functions
Abstract
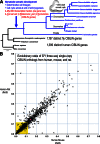
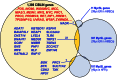
Animals use a stereotypical set of developmental genes to build body architectures of varying sizes and organizational complexity. Some genes are critical to developmental patterning, while other genes are important to physiological control of growth. However, growth regulator genes may not be as important in small-bodied "micro-metazoans" such as nematodes. Nematodes use a simplified developmental strategy of lineage-based cell fate specifications to produce an adult bilaterian body composed of a few hundreds of cells. Nematodes also lost the MYC proto-oncogenic regulator of cell proliferation. To identify additional regulators of cell proliferation that were lost with MYC, we computationally screened and determined 839 high-confidence genes that are conserved in bilaterians/lost in nematodes (CIBLIN genes). We find that 30 % of all CIBLIN genes encode transcriptional regulators of cell proliferation, epithelial-to-mesenchyme transitions, and other processes. Over 50 % of CIBLIN genes are unnamed genes in Drosophila, suggesting that there are many understudied genes. Interestingly, CIBLIN genes include many Myc synthetic lethal (MycSL) hits from recent screens. CIBLIN genes include key regulators of heparan sulfate proteoglycan (HSPG) sulfation patterns, and lysyl oxidases involved in cross-linking and modification of the extracellular matrix (ECM). These genes and others suggest the CIBLIN repertoire services critical functions in ECM remodeling and cell migration in large-bodied bilaterians. Correspondingly, CIBLIN genes are co-expressed with Myc in cancer transcriptomes, and include a preponderance of known determinants of cancer progression and tumor aggression. We propose that CIBLIN gene research can improve our understanding of regulatory control of cellular growth in metazoans.
Keywords: Apoptosis; Cancer; Cell migration; Cell proliferation; Comparative genomics; Genetic screens; Mnt; Myc; Myc synthetic lethals (MycSL genes).
Figures


Similar articles
-
Mnt takes control as key regulator of the myc/max/mxd network.Adv Cancer Res. 2007;97:61-80. doi: 10.1016/S0065-230X(06)97003-1. Adv Cancer Res. 2007. PMID: 17419941 Review.
-
The conserved Myc box 2 and Myc box 3 regions are important, but not essential, for Myc function in vivo.Gene. 2009 May 1;436(1-2):90-100. doi: 10.1016/j.gene.2009.02.009. Epub 2009 Feb 24. Gene. 2009. PMID: 19248823
-
The activities of MYC, MNT and the MAX-interactome in lymphocyte proliferation and oncogenesis.Biochim Biophys Acta. 2015 May;1849(5):554-62. doi: 10.1016/j.bbagrm.2014.04.004. Epub 2014 Apr 13. Biochim Biophys Acta. 2015. PMID: 24731854 Review.
-
Activation of transferrin receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis.Mol Cell Biol. 2006 Mar;26(6):2373-86. doi: 10.1128/MCB.26.6.2373-2386.2006. Mol Cell Biol. 2006. PMID: 16508012 Free PMC article.
-
Genomic binding and transcriptional regulation by the Drosophila Myc and Mnt transcription factors.Cold Spring Harb Symp Quant Biol. 2005;70:299-307. doi: 10.1101/sqb.2005.70.019. Cold Spring Harb Symp Quant Biol. 2005. PMID: 16869766
Cited by
-
Tango1 spatially organizes ER exit sites to control ER export.J Cell Biol. 2017 Apr 3;216(4):1035-1049. doi: 10.1083/jcb.201611088. Epub 2017 Mar 9. J Cell Biol. 2017. PMID: 28280122 Free PMC article.
-
Defective minor spliceosomes induce SMA-associated phenotypes through sensitive intron-containing neural genes in Drosophila.Nat Commun. 2020 Nov 5;11(1):5608. doi: 10.1038/s41467-020-19451-z. Nat Commun. 2020. PMID: 33154379 Free PMC article.
-
A partial genome assembly of the miniature parasitoid wasp, Megaphragma amalphitanum.PLoS One. 2019 Dec 23;14(12):e0226485. doi: 10.1371/journal.pone.0226485. eCollection 2019. PLoS One. 2019. PMID: 31869362 Free PMC article.
-
Episodic evolution of a eukaryotic NADK repertoire of ancient provenance.PLoS One. 2019 Aug 1;14(8):e0220447. doi: 10.1371/journal.pone.0220447. eCollection 2019. PLoS One. 2019. PMID: 31369599 Free PMC article.
-
A Screen for Gene Paralogies Delineating Evolutionary Branching Order of Early Metazoa.G3 (Bethesda). 2020 Feb 6;10(2):811-826. doi: 10.1534/g3.119.400951. G3 (Bethesda). 2020. PMID: 31879283 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

