Protein collapse driven against solvation free energy without H-bonds
- PMID: 26174309
- PMCID: PMC4815319
- DOI: 10.1002/pro.2749
Protein collapse driven against solvation free energy without H-bonds
Abstract
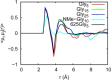
Proteins collapse and fold because intramolecular interactions and solvent entropy, which favor collapse, outweigh solute-solvent interactions that favor expansion. Since the protein backbone actively participates in protein folding and some intrinsically disordered proteins are glycine rich, oligoglycines are good models to study the protein backbone as it collapses, both during conformational changes in disordered proteins and during folding. The solvation free energies of short glycine oligomers become increasingly favorable as chain length increases. In contrast, the solubility limits of glycine oligomers decrease with increasing chain length, indicating that peptide-peptide, and potentially solvent-solvent interactions, overcome peptide-solvent interactions to favor aggregation at finite concentrations of glycine oligomers. We have recently shown that hydrogen- (H-) bonds do not contribute significantly to the concentration-based aggregation of pentaglycines but that dipole-dipole (CO) interactions between the amide groups on the backbone do. Here we demonstrate for the collapse of oligoglycines ranging in length from 15 to 25 residues similarly that H-bonds do not contribute significantly to collapse but that CO dipole interactions do. These results illustrate that some intrapeptide interactions that determine the solubility limit of short glycine oligomers are similar to those that drive the collapse of longer glycine peptides.
Keywords: intrinsically disordered proteins; molecular dynamics; oligoglycine; simulation.
© 2015 The Protein Society.
Figures



References
-
- Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181:223–230. - PubMed
-
- Flory PJ (1953) Principles of polymer chemistry. Ithaca, NY: Cornell University Press.
-
- Uversky VN, Oldfield CJ, Dunker AK (2005) Showing your ID: Intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit 18:343–384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

