Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza
- PMID: 26174967
- PMCID: PMC4502395
- DOI: 10.1038/srep11244
Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza
Abstract
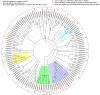
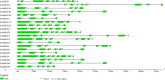
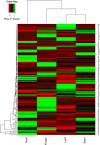
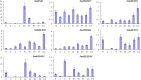
Salvia miltiorrhiza Bunge (Labiatae) is an emerging model plant for traditional medicine, and tanshinones are among the pharmacologically active constituents of this plant. Although extensive chemical and pharmaceutical studies of these compounds have been performed, studies on the basic helix-loop-helix (bHLH) transcription factors that regulate tanshinone biosynthesis are limited. In our study, 127 bHLH transcription factor genes were identified in the genome of S. miltiorrhiza, and phylogenetic analysis indicated that these SmbHLHs could be classified into 25 subfamilies. A total of 19 sequencing libraries were constructed for expression pattern analyses using RNA-Seq. Based on gene-specific expression patterns and up-regulated expression patterns in response to MeJA treatment, 7 bHLH genes were revealed as potentially involved in the regulation of tanshinone biosynthesis. Among them, the gene expression of SmbHLH37, SmbHLH74 and SmbHLH92 perfectly matches the accumulation pattern of tanshinone biosynthesis in S. miltiorrhiza. Our results provide a foundation for understanding the molecular basis and regulatory mechanisms of bHLH transcription factors in S. miltiorrhiza.
Figures

Similar articles
-
Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation.Physiol Plant. 2014 Oct;152(2):241-55. doi: 10.1111/ppl.12193. Epub 2014 May 12. Physiol Plant. 2014. PMID: 24660670
-
Effects of methyl jasmonate and salicylic acid on tanshinone production and biosynthetic gene expression in transgenic Salvia miltiorrhiza hairy roots.Biotechnol Appl Biochem. 2015 Jan-Feb;62(1):24-31. doi: 10.1002/bab.1236. Epub 2014 Jun 13. Biotechnol Appl Biochem. 2015. PMID: 24779358
-
Comprehensive transcriptome profiling of Salvia miltiorrhiza for discovery of genes associated with the biosynthesis of tanshinones and phenolic acids.Sci Rep. 2017 Sep 5;7(1):10554. doi: 10.1038/s41598-017-10215-2. Sci Rep. 2017. PMID: 28874707 Free PMC article.
-
[Salvia miltiorrhiza as medicinal model plant].Yao Xue Xue Bao. 2013 Jul;48(7):1099-106. Yao Xue Xue Bao. 2013. PMID: 24133975 Review. Chinese.
-
Tanshinone biosynthesis in Salvia miltiorrhiza and production in plant tissue cultures.Appl Microbiol Biotechnol. 2010 Sep;88(2):437-49. doi: 10.1007/s00253-010-2797-7. Epub 2010 Aug 7. Appl Microbiol Biotechnol. 2010. PMID: 20694462 Review.
Cited by
-
Genome-Wide Comprehensive Analysis of the SABATH Gene Family in Arabidopsis and Rice.Evol Bioinform Online. 2019 Jul 3;15:1176934319860864. doi: 10.1177/1176934319860864. eCollection 2019. Evol Bioinform Online. 2019. PMID: 31320793 Free PMC article.
-
The SmWRKY32-SmbHLH65/SmbHLH85 regulatory module mediates tanshinone biosynthesis in Salvia miltiorrhiza.Hortic Res. 2025 Mar 25;12(7):uhaf096. doi: 10.1093/hr/uhaf096. eCollection 2025 Jul. Hortic Res. 2025. PMID: 40391389 Free PMC article.
-
SmbHLH60 and SmMYC2 antagonistically regulate phenolic acids and anthocyanins biosynthesis in Salvia miltiorrhiza.J Adv Res. 2022 Dec;42:205-219. doi: 10.1016/j.jare.2022.02.005. Epub 2022 Feb 17. J Adv Res. 2022. PMID: 36513414 Free PMC article.
-
The bHLH transcription factor AhbHLH112 improves the drought tolerance of peanut.BMC Plant Biol. 2021 Nov 16;21(1):540. doi: 10.1186/s12870-021-03318-6. BMC Plant Biol. 2021. PMID: 34784902 Free PMC article.
-
Genome-Wide Characterization and Analysis of the bHLH Gene Family in Perilla frutescens.Int J Mol Sci. 2024 Dec 22;25(24):13717. doi: 10.3390/ijms252413717. Int J Mol Sci. 2024. PMID: 39769479 Free PMC article.
References
-
- Jaillon O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007). - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

