Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza
- PMID: 26174967
- PMCID: PMC4502395
- DOI: 10.1038/srep11244
Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza
Abstract
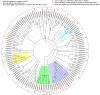
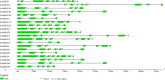
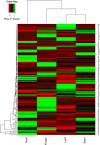
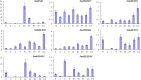
Salvia miltiorrhiza Bunge (Labiatae) is an emerging model plant for traditional medicine, and tanshinones are among the pharmacologically active constituents of this plant. Although extensive chemical and pharmaceutical studies of these compounds have been performed, studies on the basic helix-loop-helix (bHLH) transcription factors that regulate tanshinone biosynthesis are limited. In our study, 127 bHLH transcription factor genes were identified in the genome of S. miltiorrhiza, and phylogenetic analysis indicated that these SmbHLHs could be classified into 25 subfamilies. A total of 19 sequencing libraries were constructed for expression pattern analyses using RNA-Seq. Based on gene-specific expression patterns and up-regulated expression patterns in response to MeJA treatment, 7 bHLH genes were revealed as potentially involved in the regulation of tanshinone biosynthesis. Among them, the gene expression of SmbHLH37, SmbHLH74 and SmbHLH92 perfectly matches the accumulation pattern of tanshinone biosynthesis in S. miltiorrhiza. Our results provide a foundation for understanding the molecular basis and regulatory mechanisms of bHLH transcription factors in S. miltiorrhiza.
Figures

References
-
- Jaillon O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007). - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

