IMAC capture of recombinant protein from unclarified mammalian cell feed streams
- PMID: 26174988
- PMCID: PMC4737217
- DOI: 10.1002/bit.25705
IMAC capture of recombinant protein from unclarified mammalian cell feed streams
Abstract
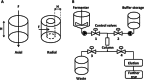
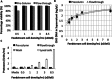
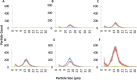
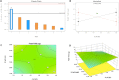
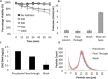
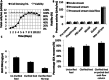
Fusion-tag affinity chromatography is a key technique in recombinant protein purification. Current methods for protein recovery from mammalian cells are hampered by the need for feed stream clarification. We have developed a method for direct capture using immobilized metal affinity chromatography (IMAC) of hexahistidine (His6) tagged proteins from unclarified mammalian cell feed streams. The process employs radial flow chromatography with 300-500 μm diameter agarose resin beads that allow free passage of cells but capture His-tagged proteins from the feed stream; circumventing expensive and cumbersome centrifugation and/or filtration steps. The method is exemplified by Chinese Hamster Ovary (CHO) cell expression and subsequent recovery of recombinant His-tagged carcinoembryonic antigen (CEA); a heavily glycosylated and clinically relevant protein. Despite operating at a high NaCl concentration necessary for IMAC binding, cells remained over 96% viable after passage through the column with host cell proteases and DNA detected at ∼ 8 U/mL and 2 ng/μL in column flow-through, respectively. Recovery of His-tagged CEA from unclarified feed yielded 71% product recovery. This work provides a basis for direct primary capture of fully glycosylated recombinant proteins from unclarified mammalian cell feed streams.
Keywords: Chinese hamster ovary cells; IMAC; downstream processing; integrated processing; radial flow chromatography; recombinant protein.
© 2015 The Authors. Biotechnology and Bioengineering Published by Wiley Periodicals, Inc.
Figures


Similar articles
-
Promiscuity of host cell proteins in the purification of histidine tagged recombinant xylanase A by IMAC procedures: A case study with a Ni2+-tacn-based IMAC system.Protein Expr Purif. 2019 Oct;162:51-61. doi: 10.1016/j.pep.2019.05.009. Epub 2019 Jun 3. Protein Expr Purif. 2019. PMID: 31170454
-
A modified IMAC method for the capture of target protein from mammalian cell culture harvest containing metal chelating species.Biotechnol Bioeng. 2012 Mar;109(3):747-53. doi: 10.1002/bit.24353. Epub 2011 Oct 28. Biotechnol Bioeng. 2012. PMID: 22012836
-
Purification of a Recombinant Polyhistidine-Tagged Glucosyltransferase Using Immobilized Metal-Affinity Chromatography (IMAC).Methods Mol Biol. 2016;1405:91-7. doi: 10.1007/978-1-4939-3393-8_9. Methods Mol Biol. 2016. PMID: 26843168
-
Enhancing the solubility of recombinant proteins in Escherichia coli by using hexahistidine-tagged maltose-binding protein as a fusion partner.Methods Mol Biol. 2011;705:259-74. doi: 10.1007/978-1-61737-967-3_16. Methods Mol Biol. 2011. PMID: 21125392 Review.
-
Enhancing aggregate reduction using anion exchange hybrid filter in an immunocytokine diabody fusion protein purification process.J Chromatogr A. 2025 Jan 4;1739:465526. doi: 10.1016/j.chroma.2024.465526. Epub 2024 Nov 16. J Chromatogr A. 2025. PMID: 39586220 Review.
Cited by
-
Purification of Membrane Proteins Overexpressed in Saccharomyces cerevisiae.Methods Mol Biol. 2022;2507:143-173. doi: 10.1007/978-1-0716-2368-8_8. Methods Mol Biol. 2022. PMID: 35773581 Free PMC article.
-
Current Advancements in Addressing Key Challenges of Therapeutic Antibody Design, Manufacture, and Formulation.Antibodies (Basel). 2019 Jun 3;8(2):36. doi: 10.3390/antib8020036. Antibodies (Basel). 2019. PMID: 31544842 Free PMC article. Review.
-
Optimization of RfxCas13d Expression in Escherichia coli Host using Response Surface Methodology.Avicenna J Med Biotechnol. 2025 Apr-Jun;17(2):122-130. doi: 10.18502/ajmb.v17i2.18563. Avicenna J Med Biotechnol. 2025. PMID: 40453911 Free PMC article.
-
RSM-based Model to Predict Optimum Fermentation Conditions for Soluble Expression of the Antibody Fragment Derived from 4D5MOC-B Humanized Mab in SHuffle™ T7 E. coli.Iran J Pharm Res. 2021 Winter;20(1):254-266. doi: 10.22037/ijpr.2020.114377.14822. Iran J Pharm Res. 2021. PMID: 34400955 Free PMC article.
References
-
- Anspach FB, Curbelo D, Hartmann R, Garke G, Deckwer W‐D. 1999. Expanded‐bed chromatography in primary protein purification. J Chromatogr A 865:129–144. - PubMed
-
- Balasundaram B, Nesbeth D, Ward JM, Keshavarz‐Moore E, Bracewell DG. 2009. Step change in the efficiency of centrifugation through cell engineering: Co‐expression of Staphylococcal nuclease to reduce the viscosity of the bioprocess feedstock. Biotechnol Bioeng 104:134–142. - PubMed
-
- Besselink T, van der Padt A, Janssen AEM, Boom RM. 2013. Are axial and radial flow chromatography different? J Chromatogr A 1271:105–114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

