Dual-colour imaging of RNAs using quencher- and fluorophore-binding aptamers
- PMID: 26175046
- PMCID: PMC4666381
- DOI: 10.1093/nar/gkv718
Dual-colour imaging of RNAs using quencher- and fluorophore-binding aptamers
Abstract
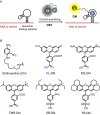
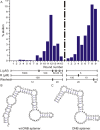
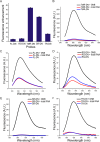
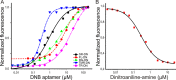
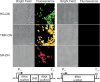
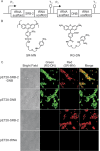
In order to gain deeper insight into the functions and dynamics of RNA in cells, the development of methods for imaging multiple RNAs simultaneously is of paramount importance. Here, we describe a modular approach to image RNA in living cells using an RNA aptamer that binds to dinitroaniline, an efficient general contact quencher. Dinitroaniline quenches the fluorescence of different fluorophores when directly conjugated to them via ethylene glycol linkers by forming a non-fluorescent intramolecular complex. Since the binding of the RNA aptamer to the quencher destroys the fluorophore-quencher complex, fluorescence increases dramatically upon binding. Using this principle, a series of fluorophores were turned into fluorescent turn-on probes by conjugating them to dinitroaniline. These probes ranged from fluorescein-dinitroaniline (green) to TexasRed-dinitroaniline (red) spanning across the visible spectrum. The dinitroaniline-binding aptamer (DNB) was generated by in vitro selection, and was found to bind all probes, leading to fluorescence increase in vitro and in living cells. When expressed in E. coli, the DNB aptamer could be labelled and visualized with different-coloured fluorophores and therefore it can be used as a genetically encoded tag to image target RNAs. Furthermore, combining contact-quenched fluorogenic probes with orthogonal DNB (the quencher-binding RNA aptamer) and SRB-2 aptamers (a fluorophore-binding RNA aptamer) allowed dual-colour imaging of two different fluorescence-enhancing RNA tags in living cells, opening new avenues for studying RNA co-localization and trafficking.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. - PubMed
-
- Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods. 2009;6:331–338. - PubMed
-
- Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. - PubMed
-
- Valencia-Burton M., McCullough R.M., Cantor C.R., Broude N.E. RNA visualization in live bacterial cells using fluorescent protein complementation. Nat. Methods. 2007;4:421–427. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

