The Saccharomyces cerevisiae Dna2 can function as a sole nuclease in the processing of Okazaki fragments in DNA replication
- PMID: 26175049
- PMCID: PMC4652754
- DOI: 10.1093/nar/gkv710
The Saccharomyces cerevisiae Dna2 can function as a sole nuclease in the processing of Okazaki fragments in DNA replication
Abstract
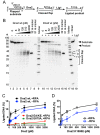
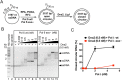
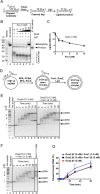
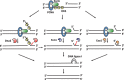
During DNA replication, synthesis of the lagging strand occurs in stretches termed Okazaki fragments. Before adjacent fragments are ligated, any flaps resulting from the displacement of the 5' DNA end of the Okazaki fragment must be cleaved. Previously, Dna2 was implicated to function upstream of flap endonuclease 1 (Fen1 or Rad27) in the processing of long flaps bound by the replication protein A (RPA). Here we show that Dna2 efficiently cleaves long DNA flaps exactly at or directly adjacent to the base. A fraction of the flaps cleaved by Dna2 can be immediately ligated. When coupled with DNA replication, the flap processing activity of Dna2 leads to a nearly complete Okazaki fragment maturation at sub-nanomolar Dna2 concentrations. Our results indicate that a subsequent nucleolytic activity of Fen1 is not required in most cases. In contrast Dna2 is completely incapable to cleave short flaps. We show that also Dna2, like Fen1, interacts with proliferating cell nuclear antigen (PCNA). We propose a model where Dna2 alone is responsible for cleaving of RPA-bound long flaps, while Fen1 or exonuclease 1 (Exo1) cleave short flaps. Our results argue that Dna2 can function in a separate, rather than in a Fen1-dependent pathway.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Significance of the dissociation of Dna2 by flap endonuclease 1 to Okazaki fragment processing in Saccharomyces cerevisiae.J Biol Chem. 2009 Mar 27;284(13):8283-91. doi: 10.1074/jbc.M809189200. Epub 2009 Jan 29. J Biol Chem. 2009. PMID: 19179330 Free PMC article.
-
Components of the secondary pathway stimulate the primary pathway of eukaryotic Okazaki fragment processing.J Biol Chem. 2010 Sep 10;285(37):28496-505. doi: 10.1074/jbc.M110.131870. Epub 2010 Jul 13. J Biol Chem. 2010. PMID: 20628185 Free PMC article.
-
Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal.J Biol Chem. 2008 Oct 10;283(41):27483-27493. doi: 10.1074/jbc.M804550200. Epub 2008 Aug 9. J Biol Chem. 2008. PMID: 18689797 Free PMC article.
-
DNA2-An Important Player in DNA Damage Response or Just Another DNA Maintenance Protein?Int J Mol Sci. 2017 Jul 18;18(7):1562. doi: 10.3390/ijms18071562. Int J Mol Sci. 2017. PMID: 28718810 Free PMC article. Review.
-
Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes.Crit Rev Biochem Mol Biol. 2010 Apr;45(2):71-96. doi: 10.3109/10409230903578593. Crit Rev Biochem Mol Biol. 2010. PMID: 20131965 Review.
Cited by
-
Direct Visualization of RNA-DNA Primer Removal from Okazaki Fragments Provides Support for Flap Cleavage and Exonucleolytic Pathways in Eukaryotic Cells.J Biol Chem. 2017 Mar 24;292(12):4777-4788. doi: 10.1074/jbc.M116.758599. Epub 2017 Feb 3. J Biol Chem. 2017. PMID: 28159842 Free PMC article.
-
Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing.PLoS Genet. 2015 Nov 6;11(11):e1005659. doi: 10.1371/journal.pgen.1005659. eCollection 2015 Nov. PLoS Genet. 2015. PMID: 26545110 Free PMC article.
-
Distinct RPA domains promote recruitment and the helicase-nuclease activities of Dna2.Nat Commun. 2021 Nov 11;12(1):6521. doi: 10.1038/s41467-021-26863-y. Nat Commun. 2021. PMID: 34764291 Free PMC article.
-
Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I.Proc Natl Acad Sci U S A. 2017 Jun 6;114(23):6010-6015. doi: 10.1073/pnas.1704845114. Epub 2017 May 22. Proc Natl Acad Sci U S A. 2017. PMID: 28533382 Free PMC article.
-
Proteasome-mediated degradation of long-range nucleases negatively regulates resection of DNA double-strand breaks.iScience. 2024 Jun 22;27(7):110373. doi: 10.1016/j.isci.2024.110373. eCollection 2024 Jul 19. iScience. 2024. PMID: 39071887 Free PMC article.
References
-
- Hubscher U., Seo Y.S. Replication of the lagging strand: a concert of at least 23 polypeptides. Mol. Cells. 2001;12:149–157. - PubMed
-
- Kang Y.H., Lee C.H., Seo Y.S. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2010;45:71–96. - PubMed
-
- Huang L., Kim Y., Turchi J.J., Bambara R.A. Structure-specific cleavage of the RNA primer from Okazaki fragments by calf thymus RNase HI. J. Biol. Chem. 1994;269:25922–25927. - PubMed
-
- Murante R.S., Rumbaugh J.A., Barnes C.J., Norton J.R., Bambara R.A. Calf RTH-1 nuclease can remove the initiator RNAs of Okazaki fragments by endonuclease activity. J. Biol. Chem. 1996;271:25888–25897. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

