Allosteric Regulation of E-Cadherin Adhesion
- PMID: 26175155
- PMCID: PMC4571897
- DOI: 10.1074/jbc.M115.657098
Allosteric Regulation of E-Cadherin Adhesion
Abstract
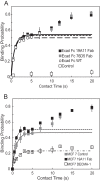
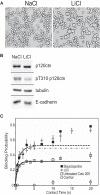
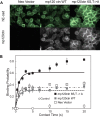
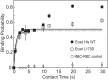
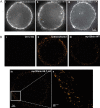
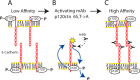
Cadherins are transmembrane adhesion proteins that maintain intercellular cohesion in all tissues, and their rapid regulation is essential for organized tissue remodeling. Despite some evidence that cadherin adhesion might be allosterically regulated, testing of this has been hindered by the difficulty of quantifying altered E-cadherin binding affinity caused by perturbations outside the ectodomain binding site. Here, measured kinetics of cadherin-mediated intercellular adhesion demonstrated quantitatively that treatment with activating, anti-E-cadherin antibodies or the dephosphorylation of a cytoplasmic binding partner, p120(ctn), increased the homophilic binding affinity of E-cadherin. Results obtained with Colo 205 cells, which express inactive E-cadherin and do not aggregate, demonstrated that four treatments, which induced Colo 205 aggregation and p120(ctn) dephosphorylation, triggered quantitatively similar increases in E-cadherin affinity. Several processes can alter cell aggregation, but these results directly demonstrated the allosteric regulation of cell surface E-cadherin by p120(ctn) dephosphorylation.
Keywords: allosteric regulation; cadherin-1 (CDH1) (epithelial cadherin) (E-cadherin); catenin; cell adhesion; kinetics.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Ciesiolka M., Delvaeye M., Van Imschoot G., Verschuere V., McCrea P., van Roy F., Vleminckx K. (2004) p120 catenin is required for morphogenetic movements involved in the formation of the eyes and the craniofacial skeleton in Xenopus. J. Cell Sci. 117, 4325–4339 - PubMed
-
- Wessel F., Winderlich M., Holm M., Frye M., Rivera-Galdos R., Vockel M., Linnepe R., Ipe U., Stadtmann A., Zarbock A., Nottebaum A. F., Vestweber D. (2014) Leukocyte extravasation and vascular permeability are each controlled in vivo by different tyrosine residues of VE-cadherin. Nat. Immunol. 15, 223–230 - PubMed
-
- Gumbiner B. M. (2005) Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 6, 622–634 - PubMed
-
- Maître J. L., Berthoumieux H., Krens S. F., Salbreux G., Jülicher F., Paluch E., Heisenberg C. P. (2012) Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science 338, 253–256 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

