Histone core phosphorylation regulates DNA accessibility
- PMID: 26175159
- PMCID: PMC4566235
- DOI: 10.1074/jbc.M115.661363
Histone core phosphorylation regulates DNA accessibility
Abstract
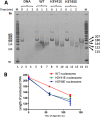
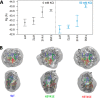
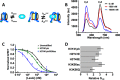
Nucleosome unwrapping dynamics provide transient access to the complexes involved in DNA transcription, repair, and replication, whereas regulation of nucleosome unwrapping modulates occupancy of these complexes. Histone H3 is phosphorylated at tyrosine 41 (H3Y41ph) and threonine 45 (H3T45ph). H3Y41ph is implicated in regulating transcription, whereas H3T45ph is involved in DNA replication and apoptosis. These modifications are located in the DNA-histone interface near where the DNA exits the nucleosome, and are thus poised to disrupt DNA-histone interactions. However, the impact of histone phosphorylation on nucleosome unwrapping and accessibility is unknown. We find that the phosphorylation mimics H3Y41E and H3T45E, and the chemically correct modification, H3Y41ph, significantly increase nucleosome unwrapping. This enhances DNA accessibility to protein binding by 3-fold. H3K56 acetylation (H3K56ac) is also located in the same DNA-histone interface and increases DNA unwrapping. H3K56ac is implicated in transcription regulation, suggesting that H3Y41ph and H3K56ac could function together. We find that the combination of H3Y41ph with H3K56ac increases DNA accessibility by over an order of magnitude. These results suggest that phosphorylation within the nucleosome DNA entry-exit region increases access to DNA binding complexes and that the combination of phosphorylation with acetylation has the potential to significantly influence DNA accessibility to transcription regulatory complexes.
Keywords: DNA accessibility; DNA-protein interaction; fluorescence resonance energy transfer (FRET); histone modification; histone post-translational modifications; nucleosome; small-angle X-ray scattering (SAXS).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 - PubMed
-
- Widom J. (1998) Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 27, 285–327 - PubMed
-
- Luger K. (2006) Dynamic nucleosomes. Chromosome Res. 14, 5–16 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

