MYC Is a Major Determinant of Mitotic Cell Fate
- PMID: 26175417
- PMCID: PMC4518499
- DOI: 10.1016/j.ccell.2015.06.001
MYC Is a Major Determinant of Mitotic Cell Fate
Abstract
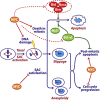
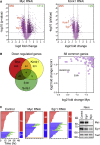
Taxol and other antimitotic agents are frontline chemotherapy agents but the mechanisms responsible for patient benefit remain unclear. Following a genome-wide siRNA screen, we identified the oncogenic transcription factor Myc as a taxol sensitizer. Using time-lapse imaging to correlate mitotic behavior with cell fate, we show that Myc sensitizes cells to mitotic blockers and agents that accelerate mitotic progression. Myc achieves this by upregulating a cluster of redundant pro-apoptotic BH3-only proteins and suppressing pro-survival Bcl-xL. Gene expression analysis of breast cancers indicates that taxane responses correlate positively with Myc and negatively with Bcl-xL. Accordingly, pharmacological inhibition of Bcl-xL restores apoptosis in Myc-deficient cells. These results open up opportunities for biomarkers and combination therapies that could enhance traditional and second-generation antimitotic agents.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







References
-
- A’Hern R.P., Jamal-Hanjani M., Szász A.M., Johnston S.R., Reis-Filho J.S., Roylance R., Swanton C. Taxane benefit in breast cancer—a role for grade and chromosomal stability. Nat. Rev. Clin. Oncol. 2013;10:357–364. - PubMed
-
- Barkett M., Gilmore T.D. Control of apoptosis by Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6910–6924. - PubMed
-
- Berns K., Horlings H.M., Hennessy B.T., Madiredjo M., Hijmans E.M., Beelen K., Linn S.C., Gonzalez-Angulo A.M., Stemke-Hale K., Hauptmann M. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

