A large-scale crop protection bioassay data set
- PMID: 26175909
- PMCID: PMC4493826
- DOI: 10.1038/sdata.2015.32
A large-scale crop protection bioassay data set
Abstract
ChEMBL is a large-scale drug discovery database containing bioactivity information primarily extracted from scientific literature. Due to the medicinal chemistry focus of the journals from which data are extracted, the data are currently of most direct value in the field of human health research. However, many of the scientific use-cases for the current data set are equally applicable in other fields, such as crop protection research: for example, identification of chemical scaffolds active against a particular target or endpoint, the de-convolution of the potential targets of a phenotypic assay, or the potential targets/pathways for safety liabilities. In order to broaden the applicability of the ChEMBL database and allow more widespread use in crop protection research, an extensive data set of bioactivity data of insecticidal, fungicidal and herbicidal compounds and assays was collated and added to the database.
Conflict of interest statement
Syngenta is a commercial organization involved in crop protection research and development.
Figures
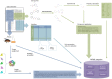
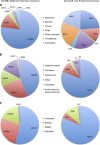
Dataset use reported in
- doi: 10.1093/nar/gkt1031
Similar articles
-
The ChEMBL database in 2017.Nucleic Acids Res. 2017 Jan 4;45(D1):D945-D954. doi: 10.1093/nar/gkw1074. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899562 Free PMC article.
-
A large-scale dataset of in vivo pharmacology assay results.Sci Data. 2018 Oct 23;5:180230. doi: 10.1038/sdata.2018.230. Sci Data. 2018. PMID: 30351302 Free PMC article.
-
Recent innovations in crop protection research.Pest Manag Sci. 2025 May;81(5):2406-2418. doi: 10.1002/ps.8441. Epub 2024 Sep 30. Pest Manag Sci. 2025. PMID: 39344983 Free PMC article. Review.
-
Oxime chemistry in crop protection.Pest Manag Sci. 2024 Sep;80(9):4163-4174. doi: 10.1002/ps.8201. Epub 2024 May 28. Pest Manag Sci. 2024. PMID: 38804722 Review.
-
ChEMBL web services: streamlining access to drug discovery data and utilities.Nucleic Acids Res. 2015 Jul 1;43(W1):W612-20. doi: 10.1093/nar/gkv352. Epub 2015 Apr 16. Nucleic Acids Res. 2015. PMID: 25883136 Free PMC article.
Cited by
-
The ChEMBL database in 2017.Nucleic Acids Res. 2017 Jan 4;45(D1):D945-D954. doi: 10.1093/nar/gkw1074. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899562 Free PMC article.
-
ChEMBL: towards direct deposition of bioassay data.Nucleic Acids Res. 2019 Jan 8;47(D1):D930-D940. doi: 10.1093/nar/gky1075. Nucleic Acids Res. 2019. PMID: 30398643 Free PMC article.
-
DRUGPATH: The Drug Gene Pathway Meta-Database.Int J Mol Sci. 2020 Apr 30;21(9):3171. doi: 10.3390/ijms21093171. Int J Mol Sci. 2020. PMID: 32365960 Free PMC article.
-
Databases and Tools to Investigate Protein-Metabolite Interactions.Methods Mol Biol. 2023;2554:231-249. doi: 10.1007/978-1-0716-2624-5_14. Methods Mol Biol. 2023. PMID: 36178629
-
Illuminating the druggable genome through patent bioactivity data.PeerJ. 2023 May 2;11:e15153. doi: 10.7717/peerj.15153. eCollection 2023. PeerJ. 2023. PMID: 37151295 Free PMC article.
References
Data Citations
-
- Gaulton A. 2014. ChEMBL. http://dx.doi.org/10.6019/CHEMBL.database.19 - DOI
References
-
- Dimova D., Stumpfe D. & Bajorath J. Systematic assessment of coordinated activity cliffs formed by kinase inhibitors and detailed characterization of activity cliff clusters and associated SAR information. Eur. J. Med. Chem. 90, 414–427 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

