Small ncRNA Expression-Profiling of Blood from Hemophilia A Patients Identifies miR-1246 as a Potential Regulator of Factor 8 Gene
- PMID: 26176629
- PMCID: PMC4503767
- DOI: 10.1371/journal.pone.0132433
Small ncRNA Expression-Profiling of Blood from Hemophilia A Patients Identifies miR-1246 as a Potential Regulator of Factor 8 Gene
Abstract
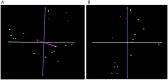
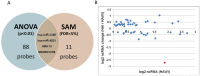
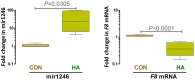
Hemophilia A (HA) is a bleeding disorder caused by deficiency of functional plasma clotting factor VIII (FVIII). Genetic mutations in the gene encoding FVIII (F8) have been extensively studied. Over a thousand different mutations have been reported in the F8 gene. These span a diverse range of mutation types, namely, missense, splice-site, deletions of single and multiple exons, inversions, etc. There is nonetheless evidence that other molecular mechanisms, in addition to mutations in the gene encoding the FVIII protein, may be involved in the pathobiology of HA. In this study, global small ncRNA expression profiling analysis of whole blood from HA patients, and controls, was performed using high-throughput ncRNA microarrays. Patients were further sub-divided into those that developed neutralizing-anti-FVIII antibodies (inhibitors) and those that did not. Selected differentially expressed ncRNAs were validated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis. We identified several ncRNAs, and among them hsa-miR-1246 was significantly up-regulated in HA patients. In addition, miR-1246 showed a six-fold higher expression in HA patients without inhibitors. We have identified an miR-1246 target site in the noncoding region of F8 mRNA and were able to confirm the suppressory role of hsa-miR-1246 on F8 expression in a stable lymphoblastoid cell line expressing FVIII. These findings suggest several testable hypotheses vis-à-vis the role of nc-RNAs in the regulation of F8 expression. These hypotheses have not been exhaustively tested in this study as they require carefully curated clinical samples.
Conflict of interest statement
Figures

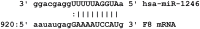
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

