Sparse whole-genome sequencing identifies two loci for major depressive disorder
- PMID: 26176920
- PMCID: PMC4522619
- DOI: 10.1038/nature14659
Sparse whole-genome sequencing identifies two loci for major depressive disorder
Erratum in
-
Author Correction: Sparse whole-genome sequencing identifies two loci for major depressive disorder.Nature. 2023 Aug;620(7976):E28. doi: 10.1038/s41586-023-06276-1. Nature. 2023. PMID: 37567929 No abstract available.
Abstract
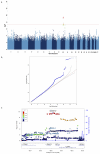
Major depressive disorder (MDD), one of the most frequently encountered forms of mental illness and a leading cause of disability worldwide, poses a major challenge to genetic analysis. To date, no robustly replicated genetic loci have been identified, despite analysis of more than 9,000 cases. Here, using low-coverage whole-genome sequencing of 5,303 Chinese women with recurrent MDD selected to reduce phenotypic heterogeneity, and 5,337 controls screened to exclude MDD, we identified, and subsequently replicated in an independent sample, two loci contributing to risk of MDD on chromosome 10: one near the SIRT1 gene (P = 2.53 × 10(-10)), the other in an intron of the LHPP gene (P = 6.45 × 10(-12)). Analysis of 4,509 cases with a severe subtype of MDD, melancholia, yielded an increased genetic signal at the SIRT1 locus. We attribute our success to the recruitment of relatively homogeneous cases with severe illness.
Figures




Comment in
-
Genetics of disease: Associations with depression.Nature. 2015 Jul 30;523(7562):539-40. doi: 10.1038/nature14635. Epub 2015 Jul 15. Nature. 2015. PMID: 26176922 No abstract available.
-
First robust genetic links to depression emerge.Nature. 2015 Jul 16;523(7560):268-9. doi: 10.1038/523268a. Nature. 2015. PMID: 26178945 No abstract available.
-
Whole-genome sequencing provides insight into the genetics of major depressive disorder.Clin Genet. 2015 Oct;88(4):340-2. doi: 10.1111/cge.12651. Clin Genet. 2015. PMID: 26260677 No abstract available.
-
China expands control over genetic data used in scientific research.Nature. 2022 May;605(7910):405. doi: 10.1038/d41586-022-01230-z. Nature. 2022. PMID: 35523869 No abstract available.
References
-
- Foley DL, et al. Genetic and environmental risk factors for depression assessed by subject-rated symptom check list versus structured clinical interview. Psychol Med. 2001;31:1413–1423. - PubMed
-
- Kendler KS, et al. Clinical indices of familial depression in the Swedish Twin Registry. Acta Psychiatrica Scandinavica. 2007;115:214–220. - PubMed

