Tandem Duplications and the Limits of Natural Selection in Drosophila yakuba and Drosophila simulans
- PMID: 26176952
- PMCID: PMC4503668
- DOI: 10.1371/journal.pone.0132184
Tandem Duplications and the Limits of Natural Selection in Drosophila yakuba and Drosophila simulans
Abstract
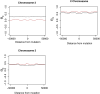
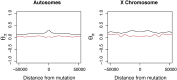
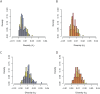
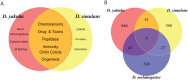
Tandem duplications are an essential source of genetic novelty, and their variation in natural populations is expected to influence adaptive walks. Here, we describe evolutionary impacts of recently-derived, segregating tandem duplications in Drosophila yakuba and Drosophila simulans. We observe an excess of duplicated genes involved in defense against pathogens, insecticide resistance, chorion development, cuticular peptides, and lipases or endopeptidases associated with the accessory glands across both species. The observed agreement is greater than expectations on chance alone, suggesting large amounts of convergence across functional categories. We document evidence of widespread selection on the D. simulans X, suggesting adaptation through duplication is common on the X. Despite the evidence for positive selection, duplicates display an excess of low frequency variants consistent with largely detrimental impacts, limiting the variation that can effectively facilitate adaptation. Standing variation for tandem duplications spans less than 25% of the genome in D. yakuba and D. simulans, indicating that evolution will be strictly limited by mutation, even in organisms with large population sizes. Effective whole gene duplication rates are low at 1.17 × 10-9 per gene per generation in D. yakuba and 6.03 × 10-10 per gene per generation in D. simulans, suggesting long wait times for new mutations on the order of thousands of years for the establishment of sweeps. Hence, in cases where adaptation depends on individual tandem duplications, evolution will be severely limited by mutation. We observe low levels of parallel recruitment of the same duplicated gene in different species, suggesting that the span of standing variation will define evolutionary outcomes in spite of convergence across gene ontologies consistent with rapidly evolving phenotypes.
Conflict of interest statement
Figures


Similar articles
-
Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans.Mol Biol Evol. 2014 Jul;31(7):1750-66. doi: 10.1093/molbev/msu124. Epub 2014 Apr 7. Mol Biol Evol. 2014. PMID: 24710518 Free PMC article.
-
Adaptive evolution of genes duplicated from the Drosophila pseudoobscura neo-X chromosome.Mol Biol Evol. 2010 Aug;27(8):1963-78. doi: 10.1093/molbev/msq085. Epub 2010 Mar 29. Mol Biol Evol. 2010. PMID: 20351054 Free PMC article.
-
Drosophila duplication hotspots are associated with late-replicating regions of the genome.PLoS Genet. 2011 Nov;7(11):e1002340. doi: 10.1371/journal.pgen.1002340. Epub 2011 Nov 3. PLoS Genet. 2011. PMID: 22072977 Free PMC article.
-
The evolution of gene duplicates.Adv Genet. 2002;46:451-83. doi: 10.1016/s0065-2660(02)46017-8. Adv Genet. 2002. PMID: 11931235 Review.
-
Role of selection in fixation of gene duplications.J Theor Biol. 2006 Mar 21;239(2):141-51. doi: 10.1016/j.jtbi.2005.08.033. Epub 2005 Oct 20. J Theor Biol. 2006. PMID: 16242725 Review.
Cited by
-
Structural variants exhibit widespread allelic heterogeneity and shape variation in complex traits.Nat Commun. 2019 Oct 25;10(1):4872. doi: 10.1038/s41467-019-12884-1. Nat Commun. 2019. PMID: 31653862 Free PMC article.
-
Gene flow mediates the role of sex chromosome meiotic drive during complex speciation.Elife. 2018 Dec 13;7:e35468. doi: 10.7554/eLife.35468. Elife. 2018. PMID: 30543325 Free PMC article.
-
Population Genomics of Two Closely Related Anhydrobiotic Midges Reveals Differences in Adaptation to Extreme Desiccation.Genome Biol Evol. 2023 Oct 6;15(10):evad169. doi: 10.1093/gbe/evad169. Genome Biol Evol. 2023. PMID: 37708413 Free PMC article.
-
Comparative population genomics of latitudinal variation in Drosophila simulans and Drosophila melanogaster.Mol Ecol. 2016 Feb;25(3):723-40. doi: 10.1111/mec.13446. Epub 2016 Jan 18. Mol Ecol. 2016. PMID: 26523848 Free PMC article.
-
Expression of tandem gene duplicates is often greater than twofold.Proc Natl Acad Sci U S A. 2016 May 24;113(21):5988-92. doi: 10.1073/pnas.1605886113. Epub 2016 May 9. Proc Natl Acad Sci U S A. 2016. PMID: 27162370 Free PMC article.
References
-
- Ohno S. Evolution by gene duplication. London: George Alien & Unwin Ltd; Berlin, Heidelberg and New York: Springer-Verlag; 1970.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

