Signal-to-Noise Ratio Measures Efficacy of Biological Computing Devices and Circuits
- PMID: 26177070
- PMCID: PMC4485182
- DOI: 10.3389/fbioe.2015.00093
Signal-to-Noise Ratio Measures Efficacy of Biological Computing Devices and Circuits
Abstract
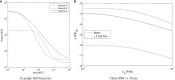
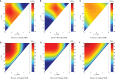
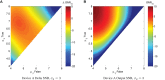
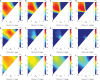
Engineering biological cells to perform computations has a broad range of important potential applications, including precision medical therapies, biosynthesis process control, and environmental sensing. Implementing predictable and effective computation, however, has been extremely difficult to date, due to a combination of poor composability of available parts and of insufficient characterization of parts and their interactions with the complex environment in which they operate. In this paper, the author argues that this situation can be improved by quantitative signal-to-noise analysis of the relationship between computational abstractions and the variation and uncertainty endemic in biological organisms. This analysis takes the form of a ΔSNRdB function for each computational device, which can be computed from measurements of a device's input/output curve and expression noise. These functions can then be combined to predict how well a circuit will implement an intended computation, as well as evaluating the general suitability of biological devices for engineering computational circuits. Applying signal-to-noise analysis to current repressor libraries shows that no library is currently sufficient for general circuit engineering, but also indicates key targets to remedy this situation and vastly improve the range of computations that can be used effectively in the implementation of biological applications.
Keywords: Boolean logic; analysis; controls; digital circuits; signals; synthetic biology.
Figures

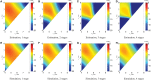
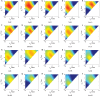
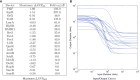
Similar articles
-
Scaling up genetic circuit design for cellular computing: advances and prospects.Nat Comput. 2018;17(4):833-853. doi: 10.1007/s11047-018-9715-9. Epub 2018 Oct 5. Nat Comput. 2018. PMID: 30524216 Free PMC article.
-
Complex cellular logic computation using ribocomputing devices.Nature. 2017 Aug 3;548(7665):117-121. doi: 10.1038/nature23271. Epub 2017 Jul 26. Nature. 2017. PMID: 28746304 Free PMC article.
-
Implementing re-configurable biological computation with distributed multicellular consortia.Nucleic Acids Res. 2022 Nov 28;50(21):12578-12595. doi: 10.1093/nar/gkac1120. Nucleic Acids Res. 2022. PMID: 36454021 Free PMC article.
-
Synthetic biology: insights into biological computation.Integr Biol (Camb). 2016 Apr 18;8(4):518-32. doi: 10.1039/c5ib00274e. Epub 2016 Apr 13. Integr Biol (Camb). 2016. PMID: 27074335 Review.
-
Engineering synthetic regulatory circuits in plants.Plant Sci. 2018 Aug;273:13-22. doi: 10.1016/j.plantsci.2018.04.005. Epub 2018 Apr 11. Plant Sci. 2018. PMID: 29907304 Review.
Cited by
-
Small molecule-inducible gene regulatory systems in mammalian cells: progress and design principles.Curr Opin Biotechnol. 2022 Dec;78:102823. doi: 10.1016/j.copbio.2022.102823. Epub 2022 Oct 27. Curr Opin Biotechnol. 2022. PMID: 36332343 Free PMC article. Review.
-
Editorial - Synthetic Biology: Engineering Complexity and Refactoring Cell Capabilities.Front Bioeng Biotechnol. 2015 Aug 21;3:120. doi: 10.3389/fbioe.2015.00120. eCollection 2015. Front Bioeng Biotechnol. 2015. PMID: 26347864 Free PMC article. No abstract available.
-
Single-cell measurement quality in bits.PLoS One. 2022 Aug 11;17(8):e0269272. doi: 10.1371/journal.pone.0269272. eCollection 2022. PLoS One. 2022. PMID: 35951522 Free PMC article.
-
Quantitative characterization of recombinase-based digitizer circuits enables predictable amplification of biological signals.Commun Biol. 2021 Jul 15;4(1):875. doi: 10.1038/s42003-021-02325-5. Commun Biol. 2021. PMID: 34267310 Free PMC article.
-
Engineering digitizer circuits for chemical and genetic screens in human cells.Nat Commun. 2021 Oct 22;12(1):6150. doi: 10.1038/s41467-021-26359-9. Nat Commun. 2021. PMID: 34686672 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

