Overfitting Bayesian Mixture Models with an Unknown Number of Components
- PMID: 26177375
- PMCID: PMC4503697
- DOI: 10.1371/journal.pone.0131739
Overfitting Bayesian Mixture Models with an Unknown Number of Components
Abstract
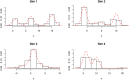
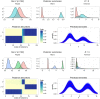
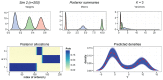
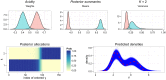
This paper proposes solutions to three issues pertaining to the estimation of finite mixture models with an unknown number of components: the non-identifiability induced by overfitting the number of components, the mixing limitations of standard Markov Chain Monte Carlo (MCMC) sampling techniques, and the related label switching problem. An overfitting approach is used to estimate the number of components in a finite mixture model via a Zmix algorithm. Zmix provides a bridge between multidimensional samplers and test based estimation methods, whereby priors are chosen to encourage extra groups to have weights approaching zero. MCMC sampling is made possible by the implementation of prior parallel tempering, an extension of parallel tempering. Zmix can accurately estimate the number of components, posterior parameter estimates and allocation probabilities given a sufficiently large sample size. The results will reflect uncertainty in the final model and will report the range of possible candidate models and their respective estimated probabilities from a single run. Label switching is resolved with a computationally light-weight method, Zswitch, developed for overfitted mixtures by exploiting the intuitiveness of allocation-based relabelling algorithms and the precision of label-invariant loss functions. Four simulation studies are included to illustrate Zmix and Zswitch, as well as three case studies from the literature. All methods are available as part of the R package Zmix, which can currently be applied to univariate Gaussian mixture models.
Conflict of interest statement
Figures

Similar articles
-
Part 2. Development of Enhanced Statistical Methods for Assessing Health Effects Associated with an Unknown Number of Major Sources of Multiple Air Pollutants.Res Rep Health Eff Inst. 2015 Jun;(183 Pt 1-2):51-113. Res Rep Health Eff Inst. 2015. PMID: 26333239
-
Allocation Variable-Based Probabilistic Algorithm to Deal with Label Switching Problem in Bayesian Mixture Models.PLoS One. 2015 Oct 12;10(10):e0138899. doi: 10.1371/journal.pone.0138899. eCollection 2015. PLoS One. 2015. PMID: 26458185 Free PMC article.
-
A probabilistic solution to the MEG inverse problem via MCMC methods: the reversible jump and parallel tempering algorithms.IEEE Trans Biomed Eng. 2001 May;48(5):533-42. doi: 10.1109/10.918592. IEEE Trans Biomed Eng. 2001. PMID: 11341527
-
Bayesian mixture models of variable dimension for image segmentation.Comput Methods Programs Biomed. 2009 Apr;94(1):1-14. doi: 10.1016/j.cmpb.2008.05.010. Epub 2008 Nov 25. Comput Methods Programs Biomed. 2009. PMID: 19036468
-
A primer on Bayesian inference for biophysical systems.Biophys J. 2015 May 5;108(9):2103-13. doi: 10.1016/j.bpj.2015.03.042. Biophys J. 2015. PMID: 25954869 Free PMC article. Review.
Cited by
-
Disentangling Qualitatively Different Faking Strategies in High-Stakes Personality Assessments: A Mixture Extension of the Multidimensional Nominal Response Model.Educ Psychol Meas. 2025 Jul 29:00131644251341843. doi: 10.1177/00131644251341843. Online ahead of print. Educ Psychol Meas. 2025. PMID: 40756699 Free PMC article.
-
Identifying dietary consumption patterns from survey data: a Bayesian nonparametric latent class model.J R Stat Soc Ser A Stat Soc. 2023 Dec 12;187(2):496-512. doi: 10.1093/jrsssa/qnad135. eCollection 2024 Apr. J R Stat Soc Ser A Stat Soc. 2023. PMID: 38617597 Free PMC article.
-
PyClone-VI: scalable inference of clonal population structures using whole genome data.BMC Bioinformatics. 2020 Dec 10;21(1):571. doi: 10.1186/s12859-020-03919-2. BMC Bioinformatics. 2020. PMID: 33302872 Free PMC article.
-
Robust Clustering with Subpopulation-specific Deviations.J Am Stat Assoc. 2020;115(530):521-537. doi: 10.1080/01621459.2019.1611583. Epub 2019 Jun 19. J Am Stat Assoc. 2020. PMID: 32952235 Free PMC article.
-
Empirically Derived Dietary Patterns Using Robust Profile Clustering in the Hispanic Community Health Study/Study of Latinos.J Nutr. 2020 Oct 12;150(10):2825-2834. doi: 10.1093/jn/nxaa208. J Nutr. 2020. PMID: 32710754 Free PMC article.
References
-
- Fruhwirth-Schnatter SI. Finite mixture and Markov switching models. 1st ed Springer; 2006. PLOS 20/232.
-
- White N, Johnson H, Silburn P, Mellick G, Dissanayaka N, Mengersen K. Probabilistic subgroup identification using Bayesian finite mixture modelling: A case study in Parkinson’s disease phenotype identification. Statistical methods in medical research. 2010 Dec;. - PubMed
-
- Heckman JJ, Taber CR. Econometric mixture models and more general models for unobservables in duration analysis Statistical Methods in Medical Research. 1994;3(3):279–299. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

