miR-34 and p53: New Insights into a Complex Functional Relationship
- PMID: 26177460
- PMCID: PMC4503669
- DOI: 10.1371/journal.pone.0132767
miR-34 and p53: New Insights into a Complex Functional Relationship
Abstract
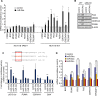
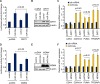
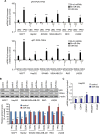
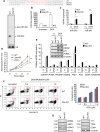
miR-34, a tumor suppressor miRNA family transcriptionally activated by p53, is considered a critical mediator of p53 function. However, knockout of the mouse miR-34 family has little or no effect on the p53 response. The relative contribution of different miR-34 family members to p53 function or how much p53 relies on miR-34 in human cells is unclear. Here we show that miR-34a has a complex effect on the p53 response in human cells. In HCT116 cells miR-34a overexpression enhances p53 transcriptional activity, but the closely related family members, miR-34b and miR-34c, even when over-expressed, have little effect. Both TP53 itself and MDM4, a strong p53 transactivation inhibitor, are direct targets of miR-34a. The genes regulated by miR-34a also include four other post-translational inhibitors of p53. miR-34a overexpression leads to variable effects on p53 levels in p53-sufficient human cancer cell lines. In HCT116, miR-34a overexpression increases p53 protein levels and stability. About a quarter of all mRNAs that participate in the human p53 network bind to biotinylated miR-34a, suggesting that many are direct miR-34a targets. However, only about a fifth of the mRNAs that bind to miR-34a also bind to miR-34b or miR-34c. Two human cell lines knocked out for miR-34a have unimpaired p53-mediated responses to genotoxic stress, like mouse cells. The complex positive and negative effects of miR-34 on the p53 network suggest that rather than simply promoting the p53 response, miR-34a might act at a systems level to stabilize the robustness of the p53 response to genotoxic stress.
Conflict of interest statement
Figures




Similar articles
-
MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame.PLoS One. 2012;7(8):e42034. doi: 10.1371/journal.pone.0042034. Epub 2012 Aug 1. PLoS One. 2012. PMID: 22870278 Free PMC article.
-
CRISPR/Cas9-mediated inactivation of miR-34a and miR-34b/c in HCT116 colorectal cancer cells: comprehensive characterization after exposure to 5-FU reveals EMT and autophagy as key processes regulated by miR-34.Cell Death Differ. 2023 Aug;30(8):2017-2034. doi: 10.1038/s41418-023-01193-2. Epub 2023 Jul 24. Cell Death Differ. 2023. PMID: 37488217 Free PMC article.
-
c-Jun N-terminal kinase 1/c-Jun activation of the p53/microRNA 34a/sirtuin 1 pathway contributes to apoptosis induced by deoxycholic acid in rat liver.Mol Cell Biol. 2014 Mar;34(6):1100-20. doi: 10.1128/MCB.00420-13. Epub 2014 Jan 13. Mol Cell Biol. 2014. PMID: 24421392 Free PMC article.
-
Modeling miRNA regulation in cancer signaling systems: miR-34a regulation of the p53/Sirt1 signaling module.Methods Mol Biol. 2012;880:87-108. doi: 10.1007/978-1-61779-833-7_6. Methods Mol Biol. 2012. PMID: 23361983 Review.
-
The guardian's little helper: microRNAs in the p53 tumor suppressor network.Cancer Res. 2007 Dec 1;67(23):11099-101. doi: 10.1158/0008-5472.CAN-07-2672. Cancer Res. 2007. PMID: 18056431 Review.
Cited by
-
New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies.Int J Mol Sci. 2016 Apr 27;17(5):627. doi: 10.3390/ijms17050627. Int J Mol Sci. 2016. PMID: 27128908 Free PMC article. Review.
-
MicroRNA Methylome Signature and Their Functional Roles in Colorectal Cancer Diagnosis, Prognosis, and Chemoresistance.Int J Mol Sci. 2022 Jun 30;23(13):7281. doi: 10.3390/ijms23137281. Int J Mol Sci. 2022. PMID: 35806286 Free PMC article. Review.
-
Context-Dependent Regulation of Gene Expression by Non-Canonical Small RNAs.Noncoding RNA. 2022 Apr 29;8(3):29. doi: 10.3390/ncrna8030029. Noncoding RNA. 2022. PMID: 35645336 Free PMC article. Review.
-
The Role of miRNAs in the Development, Proliferation, and Progression of Endometrial Cancer.Int J Mol Sci. 2023 Jul 15;24(14):11489. doi: 10.3390/ijms241411489. Int J Mol Sci. 2023. PMID: 37511248 Free PMC article. Review.
-
Evaluation of long-chain acyl-coenzyme A synthetase 4 (ACSL4) expression in human breast cancer.Res Pharm Sci. 2020 Feb 20;15(1):48-56. doi: 10.4103/1735-5362.278714. eCollection 2020 Feb. Res Pharm Sci. 2020. PMID: 32180816 Free PMC article.
References
-
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17(15):1298–307. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

