Heterogeneity among Homologs of Cutinase-Like Protein Cut5 in Mycobacteria
- PMID: 26177502
- PMCID: PMC4503659
- DOI: 10.1371/journal.pone.0133186
Heterogeneity among Homologs of Cutinase-Like Protein Cut5 in Mycobacteria
Abstract
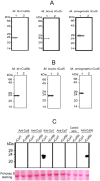
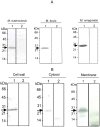
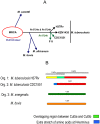
The study of genomic variability within various pathogenic and non-pathogenic strains of mycobacteria provides insight into their evolution and pathogenesis. The mycobacterial genome encodes seven cutinase-like proteins and each one of these exhibit distinct characteristics. We describe the presence of Cut5, a member of the cutinase family, in mycobacteria and the existence of a unique genomic arrangement in the cut5 gene of M. tuberculosis (Mtb) strains. A single nucleotide (T) insertion is observed in the cut5 gene, which is specific for Mtb strains. Using in silico analysis and RT-PCR, we demonstrate the transcription of Rv3724/cut5 as Rv3724a/cut5a and Rv3724b/cut5b in Mtb H37Rv and as full length cut5 in M. bovis. Cut5b protein of Mtb H37Rv (MtbCut5b) was found to be antigenically similar to its homologs in M. bovis and M. smegmatis, without any observed cross-reactivity with other Mtb cutinases. Also, the presence of Cut5b in Mtb and its homologs in M. bovis and M. smegmatis were confirmed by western blotting using antibodies raised against recombinant Cut5b. In Mtb H37Rv, Cut5b was found to be localized in the cell wall, cytosol and membrane fractions. We also report the vast prevalence of Cut5 homologs in pathogenic and non pathogenic species of mycobacteria. In silico analysis revealed that this protein has three possible organizations in mycobacteria. Also, a single nucleotide (T) insertion in Mtb strains and varied genomic arrangements within mycobacterial species make Rv3724/Cut5 a potential candidate that can be exploited as a biomarker in Mtb infection.
Conflict of interest statement
Figures




Similar articles
-
Purification and characterization of mycobacterial phospholipase A: an activity associated with mycobacterial cutinase.J Bacteriol. 2007 Jun;189(11):4153-60. doi: 10.1128/JB.01909-06. Epub 2007 Apr 6. J Bacteriol. 2007. PMID: 17416658 Free PMC article.
-
Mycobacterium tuberculosis mammalian cell entry operon (mce) homologs in Mycobacterium other than tuberculosis (MOTT).FEMS Immunol Med Microbiol. 2002 Jun 3;33(2):125-32. doi: 10.1111/j.1574-695X.2002.tb00581.x. FEMS Immunol Med Microbiol. 2002. PMID: 12052567
-
Use of the hupB gene encoding a histone-like protein of Mycobacterium tuberculosis as a target for detection and differentiation of M. tuberculosis and M. bovis.J Clin Microbiol. 2004 Jun;42(6):2724-32. doi: 10.1128/JCM.42.6.2724-2732.2004. J Clin Microbiol. 2004. PMID: 15184459 Free PMC article.
-
Roles of Triolein and Lipolytic Protein in the Pathogenesis and Survival of Mycobacterium tuberculosis: a Novel Therapeutic Approach.Appl Biochem Biotechnol. 2016 Apr;178(7):1377-89. doi: 10.1007/s12010-015-1953-z. Epub 2015 Dec 17. Appl Biochem Biotechnol. 2016. PMID: 26679705 Review.
-
Role of TlyA in the Biology of Uncultivable Mycobacteria.Comb Chem High Throughput Screen. 2022;25(10):1587-1594. doi: 10.2174/1386207325666220111150923. Comb Chem High Throughput Screen. 2022. PMID: 35021968 Review.
Cited by
-
Expression of a Cutinase of Moniliophthora roreri with Polyester and PET-Plastic Residues Degradation Activity.Microbiol Spectr. 2021 Dec 22;9(3):e0097621. doi: 10.1128/Spectrum.00976-21. Epub 2021 Nov 3. Microbiol Spectr. 2021. PMID: 34730414 Free PMC article.
-
Roles of Lipolytic enzymes in Mycobacterium tuberculosis pathogenesis.Front Microbiol. 2024 Jan 29;15:1329715. doi: 10.3389/fmicb.2024.1329715. eCollection 2024. Front Microbiol. 2024. PMID: 38357346 Free PMC article. Review.
References
-
- Chegou NN, Black GF, Loxton AG, Stanley K, Essone PN, Klein MR, et al. Potential of novel Mycobacterium tuberculosis infection phase-dependent antigens in the diagnosis of TB disease in a high burden setting. BMC infectious diseases. 2012;12:10 Epub 2012/01/21. 10.1186/1471-2334-12-10 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

