PpCBF3 from Cold-Tolerant Kentucky Bluegrass Involved in Freezing Tolerance Associated with Up-Regulation of Cold-Related Genes in Transgenic Arabidopsis thaliana
- PMID: 26177510
- PMCID: PMC4503346
- DOI: 10.1371/journal.pone.0132928
PpCBF3 from Cold-Tolerant Kentucky Bluegrass Involved in Freezing Tolerance Associated with Up-Regulation of Cold-Related Genes in Transgenic Arabidopsis thaliana
Abstract
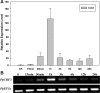
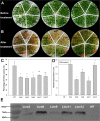
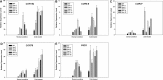
Dehydration-Responsive Element Binding proteins (DREB)/C-repeat (CRT) Binding Factors (CBF) have been identified as transcriptional activators during plant responses to cold stress. The objective of this study was to determine the physiological roles of a CBF gene isolated from a cold-tolerant perennial grass species, Kentucky bluegrass (Poa pratensis L.), which designated as PpCBF3, in regulating plant tolerance to freezing stress. Transient transformation of Arabidopsis thaliana mesophyll protoplast with PpCBF3-eGFP fused protein showed that PpCBF3 was localized to the nucleus. RT-PCR analysis showed that PpCBF3 was specifically induced by cold stress (4°C) but not by drought stress [induced by 20% polyethylene glycol 6000 solution (PEG-6000)] or salt stress (150 mM NaCl). Transgenic Arabidopsis overexpressing PpCBF3 showed significant improvement in freezing (-20°C) tolerance demonstrated by a lower percentage of chlorotic leaves, lower cellular electrolyte leakage (EL) and H2O2 and O2.- content, and higher chlorophyll content and photochemical efficiency compared to the wild type. Relative mRNA expression level analysis by qRT-PCR indicated that the improved freezing tolerance of transgenic Arabidopsis plants overexpressing PpCBF3 was conferred by sustained activation of downstream cold responsive (COR) genes. Other interesting phenotypic changes in the PpCBF3-transgenic Arabidopsis plants included late flowering and slow growth or 'dwarfism', both of which are desirable phenotypic traits for perennial turfgrasses. Therefore, PpCBF3 has potential to be used in genetic engineering for improvement of turfgrass freezing tolerance and other desirable traits.
Conflict of interest statement
Figures



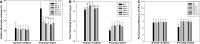


Similar articles
-
Ectopic overexpression of SsCBF1, a CRT/DRE-binding factor from the nightshade plant Solanum lycopersicoides, confers freezing and salt tolerance in transgenic Arabidopsis.PLoS One. 2013 Jun 3;8(6):e61810. doi: 10.1371/journal.pone.0061810. Print 2014. PLoS One. 2013. PMID: 23755095 Free PMC article.
-
Overexpression of a novel cold-responsive transcript factor LcFIN1 from sheepgrass enhances tolerance to low temperature stress in transgenic plants.Plant Biotechnol J. 2016 Mar;14(3):861-74. doi: 10.1111/pbi.12435. Epub 2015 Aug 3. Plant Biotechnol J. 2016. PMID: 26234381 Free PMC article.
-
Isolation and functional characterization of a cold responsive phosphatidylinositol transfer-associated protein, ZmSEC14p, from maize (Zea may L.).Plant Cell Rep. 2016 Aug;35(8):1671-86. doi: 10.1007/s00299-016-1980-4. Epub 2016 Apr 9. Plant Cell Rep. 2016. PMID: 27061906
-
[Arabidopsis CBF1 in plant tolerance to low temperature and drought stresses].Yi Chuan. 2004 May;26(3):394-8. Yi Chuan. 2004. PMID: 15640027 Review. Chinese.
-
Natural Variation in Freezing Tolerance and Cold Acclimation Response in Arabidopsis thaliana and Related Species.Adv Exp Med Biol. 2018;1081:81-98. doi: 10.1007/978-981-13-1244-1_5. Adv Exp Med Biol. 2018. PMID: 30288705 Review.
Cited by
-
SVALKA: A Long Noncoding Cis-Natural Antisense RNA That Plays a Role in the Regulation of the Cold Response of Arabidopsis thaliana.Noncoding RNA. 2024 Nov 28;10(6):59. doi: 10.3390/ncrna10060059. Noncoding RNA. 2024. PMID: 39728604 Free PMC article. Review.
-
Transgenic crops for the agricultural improvement in Pakistan: a perspective of environmental stresses and the current status of genetically modified crops.GM Crops Food. 2020;11(1):1-29. doi: 10.1080/21645698.2019.1680078. Epub 2019 Nov 3. GM Crops Food. 2020. PMID: 31679447 Free PMC article. Review.
-
Coping with the cold: unveiling cryoprotectants, molecular signaling pathways, and strategies for cold stress resilience.Front Plant Sci. 2023 Aug 15;14:1246093. doi: 10.3389/fpls.2023.1246093. eCollection 2023. Front Plant Sci. 2023. PMID: 37649996 Free PMC article. Review.
-
Meta-Analysis of the Effect of Overexpression of Dehydration-Responsive Element Binding Family Genes on Temperature Stress Tolerance and Related Responses.Front Plant Sci. 2018 May 29;9:713. doi: 10.3389/fpls.2018.00713. eCollection 2018. Front Plant Sci. 2018. PMID: 29896212 Free PMC article.
-
ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress.Int J Mol Sci. 2022 Jan 28;23(3):1549. doi: 10.3390/ijms23031549. Int J Mol Sci. 2022. PMID: 35163471 Free PMC article. Review.
References
-
- Kourosh V, Charles L. Abiotic Stress-Plant Responses and Applications in Agriculture In: Hasanuzzaman M, Nahar K, Fujita M, editors. Extreme Temperature Responses, Oxidative Stress and Antioxidant Defense in Plants. INTECH Open Access Publisher; 2013. pp. 169–205.
-
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998; 10: 1391–1406. - PMC - PubMed
-
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997; 94: 1035–1040. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

