Integrated sequence and expression analysis of ovarian cancer structural variants underscores the importance of gene fusion regulation
- PMID: 26177635
- PMCID: PMC4504069
- DOI: 10.1186/s12920-015-0118-9
Integrated sequence and expression analysis of ovarian cancer structural variants underscores the importance of gene fusion regulation
Abstract
Background: Genomic rearrangements or structural variants (SVs) are one of the most common classes of mutations in cancer.
Methods: An integrated DNA sequencing and transcriptional profiling (RNA sequence and microarray gene expression data) analysis was performed on six ovarian cancer patient samples. Matched sets of control (whole blood) samples from these same patients were used to distinguish cancer SVs of germline origin from those arising somatically in the cancer cell lineage.
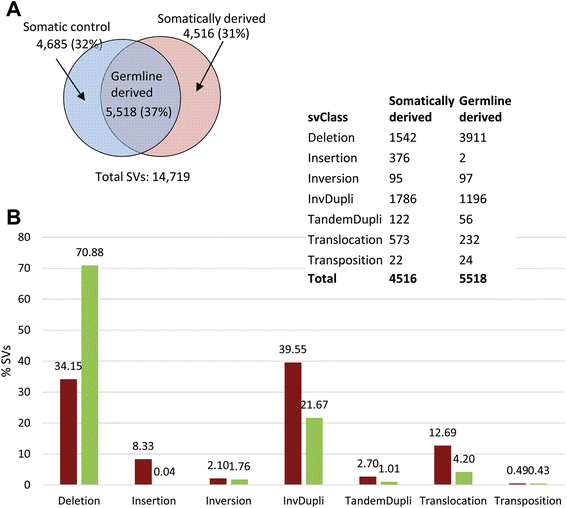
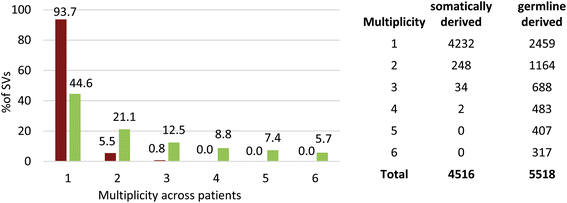
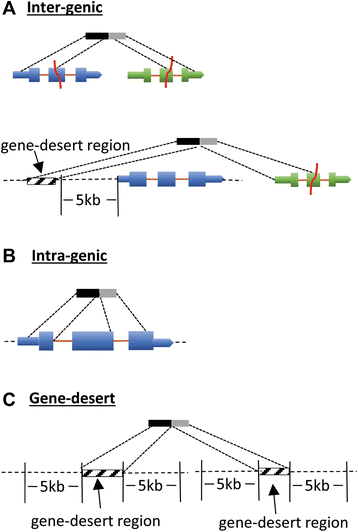
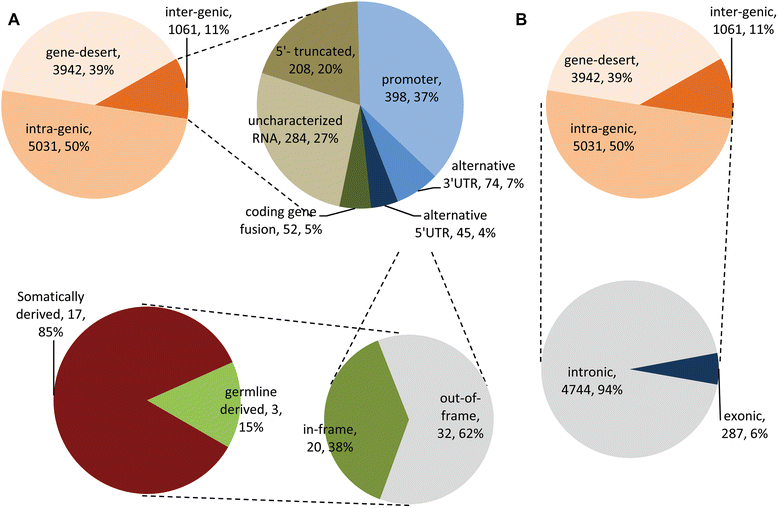
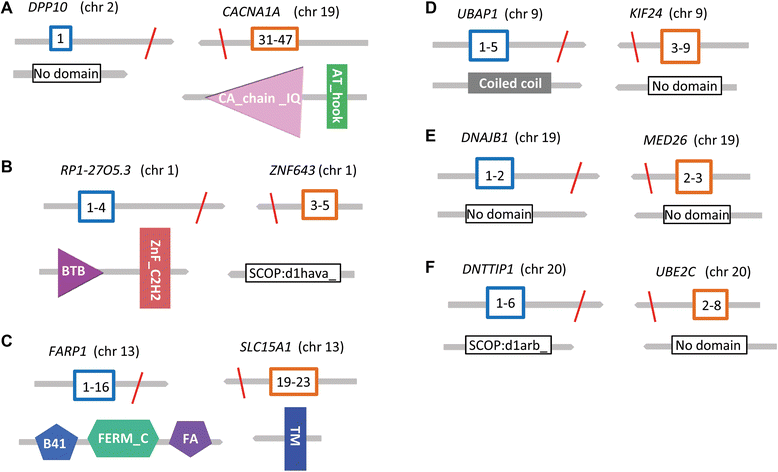
Results: We detected 10,034 ovarian cancer SVs (5518 germline derived; 4516 somatically derived) at base-pair level resolution. Only 11 % of these variants were shown to have the potential to form gene fusions and, of these, less than 20 % were detected at the transcriptional level.
Conclusions: Collectively our results are consistent with the view that gene fusions and other SVs can be significant factors in the onset and progression of ovarian cancer. The results further indicate that it may not only be the occurrence of these variants in cancer but their regulation that contributes to their biological and clinical significance.
Figures
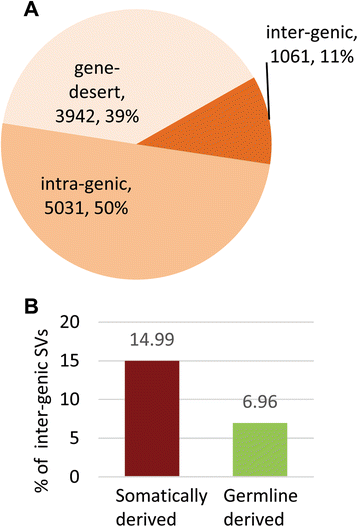
References
-
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, Ye K, Allpaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff OB, Spiridou A, Stebbins LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell P, Bentley DR, Futreal PA, Stratton MR. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. - DOI - PMC - PubMed
-
- Edwards PA. Fusion genes and chromosome translocations in the common epithelial cancers. J Path. 2010;220:244–254. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

