Structural and Functional Characterization of a Lytic Polysaccharide Monooxygenase with Broad Substrate Specificity
- PMID: 26178376
- PMCID: PMC4645601
- DOI: 10.1074/jbc.M115.660183
Structural and Functional Characterization of a Lytic Polysaccharide Monooxygenase with Broad Substrate Specificity
Abstract
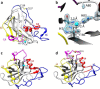
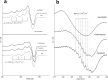
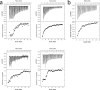
The recently discovered lytic polysaccharide monooxygenases (LPMOs) carry out oxidative cleavage of polysaccharides and are of major importance for efficient processing of biomass. NcLPMO9C from Neurospora crassa acts both on cellulose and on non-cellulose β-glucans, including cellodextrins and xyloglucan. The crystal structure of the catalytic domain of NcLPMO9C revealed an extended, highly polar substrate-binding surface well suited to interact with a variety of sugar substrates. The ability of NcLPMO9C to act on soluble substrates was exploited to study enzyme-substrate interactions. EPR studies demonstrated that the Cu(2+) center environment is altered upon substrate binding, whereas isothermal titration calorimetry studies revealed binding affinities in the low micromolar range for polymeric substrates that are due in part to the presence of a carbohydrate-binding module (CBM1). Importantly, the novel structure of NcLPMO9C enabled a comparative study, revealing that the oxidative regioselectivity of LPMO9s (C1, C4, or both) correlates with distinct structural features of the copper coordination sphere. In strictly C1-oxidizing LPMO9s, access to the solvent-facing axial coordination position is restricted by a conserved tyrosine residue, whereas access to this same position seems unrestricted in C4-oxidizing LPMO9s. LPMO9s known to produce a mixture of C1- and C4-oxidized products show an intermediate situation.
Keywords: biodegradation; bioenergy; copper monooxygenase; crystal structure; electron paramagnetic resonance (EPR); isothermal titration calorimetry (ITC).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

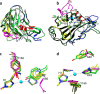


References
-
- Harris P. V., Welner D., McFarland K. C., Re E., Navarro Poulsen J. C., Brown K., Salbo R., Ding H., Vlasenko E., Merino S., Xu F., Cherry J., Larsen S., Lo Leggio L. (2010) Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49, 3305–3316 - PubMed
-
- Phillips C. M., Beeson W. T., Cate J. H., Marletta M. A. (2011) Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6, 1399–1406 - PubMed
-
- Quinlan R. J., Sweeney M. D., Lo Leggio L., Otten H., Poulsen J. C., Johansen K. S., Krogh K. B., Jørgensen C. I., Tovborg M., Anthonsen A., Tryfona T., Walter C. P., Dupree P., Xu F., Davies G. J., Walton P. H. (2011) Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U.S.A. 108, 15079–15084 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

