The de novo genome assembly and annotation of a female domestic dromedary of North African origin
- PMID: 26178449
- PMCID: PMC4973839
- DOI: 10.1111/1755-0998.12443
The de novo genome assembly and annotation of a female domestic dromedary of North African origin
Abstract
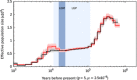
The single-humped dromedary (Camelus dromedarius) is the most numerous and widespread of domestic camel species and is a significant source of meat, milk, wool, transportation and sport for millions of people. Dromedaries are particularly well adapted to hot, desert conditions and harbour a variety of biological and physiological characteristics with evolutionary, economic and medical importance. To understand the genetic basis of these traits, an extensive resource of genomic variation is required. In this study, we assembled at 65× coverage, a 2.06 Gb draft genome of a female dromedary whose ancestry can be traced to an isolated population from the Canary Islands. We annotated 21,167 protein-coding genes and estimated ~33.7% of the genome to be repetitive. A comparison with the recently published draft genome of an Arabian dromedary resulted in 1.91 Gb of aligned sequence with a divergence of 0.095%. An evaluation of our genome with the reference revealed that our assembly contains more error-free bases (91.2%) and fewer scaffolding errors. We identified ~1.4 million single-nucleotide polymorphisms with a mean density of 0.71 × 10(-3) per base. An analysis of demographic history indicated that changes in effective population size corresponded with recent glacial epochs. Our de novo assembly provides a useful resource of genomic variation for future studies of the camel's adaptations to arid environments and economically important traits. Furthermore, these results suggest that draft genome assemblies constructed with only two differently sized sequencing libraries can be comparable to those sequenced using additional library sizes, highlighting that additional resources might be better placed in technologies alternative to short-read sequencing to physically anchor scaffolds to genome maps.
Keywords: Camelus dromedarius; adaptation; demography; domestication; next-generation sequencing.
© 2015 The Authors. Molecular Ecology Resources Published by John Wiley & Sons Ltd.
Figures
References
-
- Agrawal RP, Jain S, Shah S, et al (2011) Effect of camel milk on glycemic control and insulin requirement in patients with type 1 diabetes: 2‐years randomized controlled trial. European Journal of Clinical Nutrition, 65, 1048–1052. - PubMed
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

