BCR/ABL1 and BCR are under the transcriptional control of the MYC oncogene
- PMID: 26179066
- PMCID: PMC4504180
- DOI: 10.1186/s12943-015-0407-0
BCR/ABL1 and BCR are under the transcriptional control of the MYC oncogene
Abstract
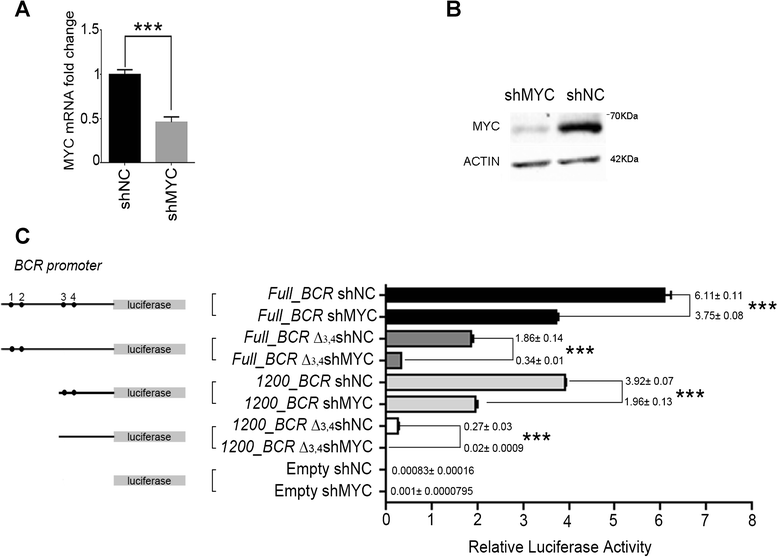
Background: Chronic Myeloid Leukaemia (CML) is caused by the BCR/ABL1 fusion gene. Both the presence and the levels of BCR/ABL1 expression seem to be critical for CML progression from chronic phase (CP) to blast crisis (BC). After the oncogenic translocation, the BCR/ABL1 gene is under the transcriptional control of BCR promoter but the molecular mechanisms involved in the regulation of oncogene expression are mostly unknown.
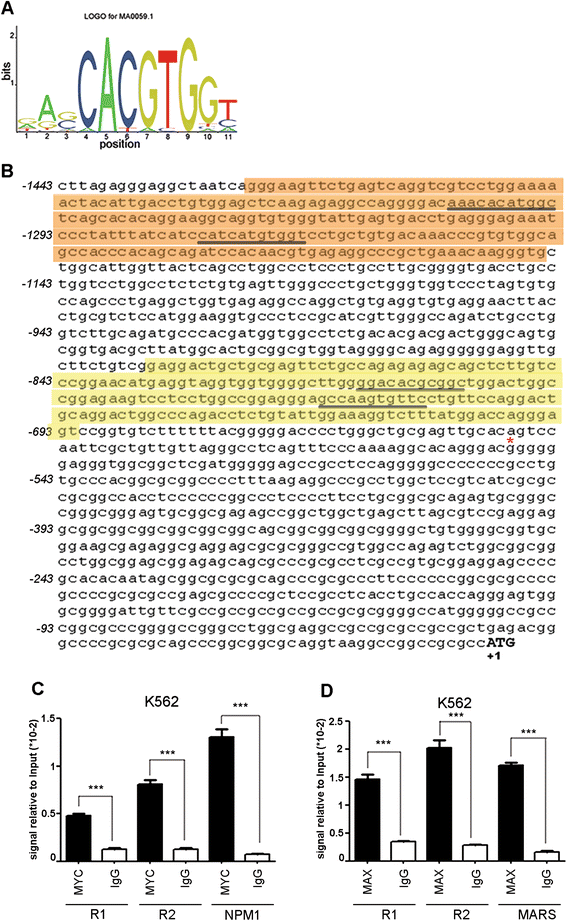
Methods: A region of 1443bp of the functional BCR promoter was studied for transcription factor binding sites through in-silico analysis and Chromatin Immunoprecipitation experiments. BCR and BCR/ABL1 expression levels were analysed in CML cell lines after over-expression or silencing of MYC transcription factor. A luciferase reporter assay was used to confirm its activity on BCR promoter.
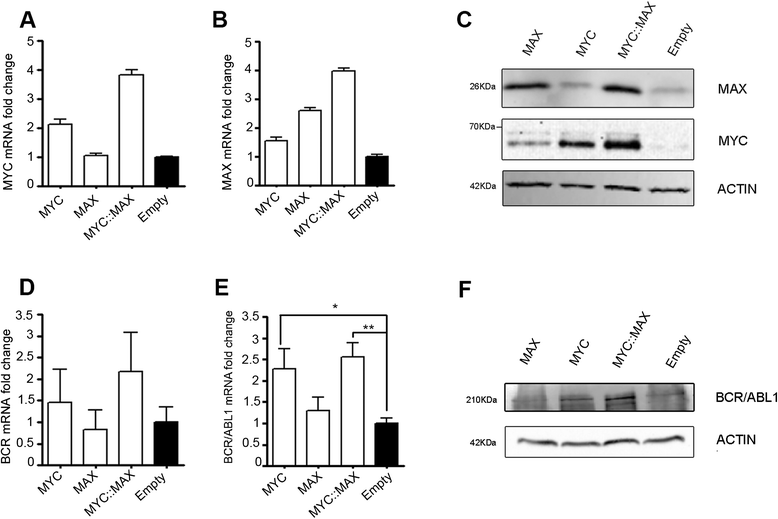
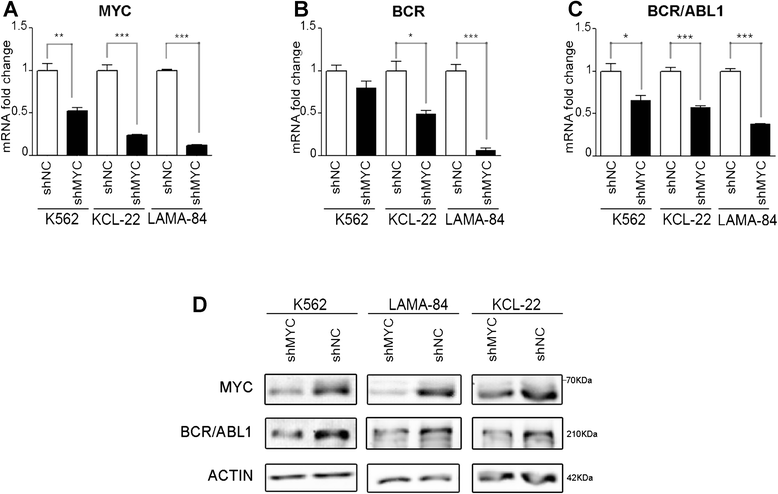
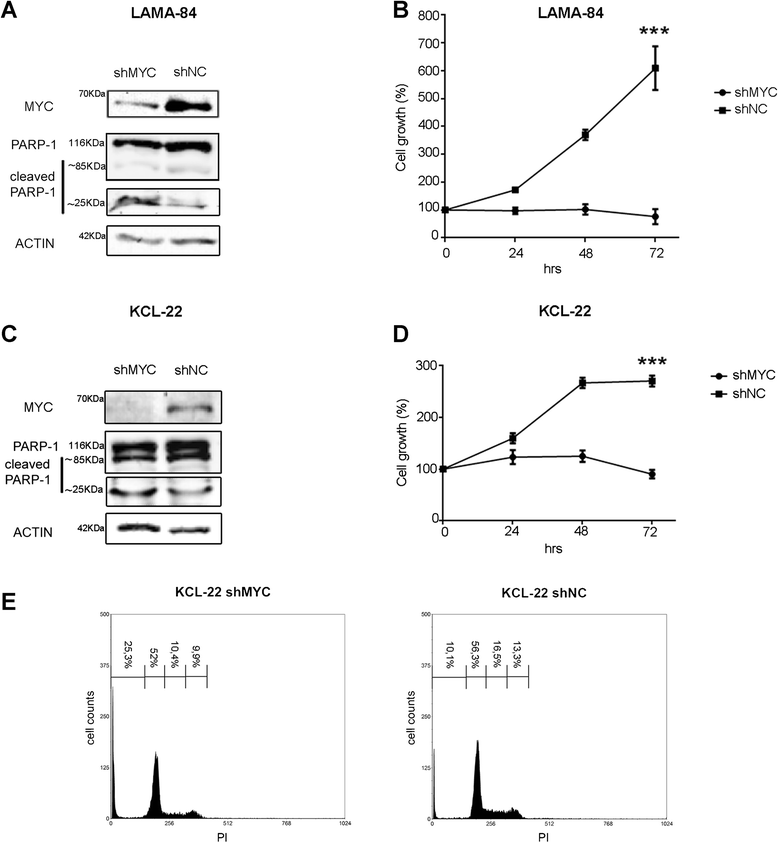
Results: In the present study we demonstrate that MYC and its partner MAX bind to the BCR promoter, leading to up-regulation of BCR and BCR/ABL1 at both transcriptional and protein levels. Accordingly, silencing of MYC expression in various BCR/ABL1 positive cell lines causes significant downregulation of BCR and BCR/ABL1, which consequently leads to decreased proliferation and induction of cell death.
Conclusions: Here we describe a regulatory pathway modulating BCR and BCR/ABL1 expression, showing that the BCR promoter is under the transcriptional control of the MYC/MAX heterodimer. Since MYC is frequently over-expressed in BC, this phenomenon could play a critical role in BCR/ABL1 up-regulation and blast aggressiveness acquired during CML evolution.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

