The cell cycle regulator 14-3-3σ opposes and reverses cancer metabolic reprogramming
- PMID: 26179207
- PMCID: PMC4507299
- DOI: 10.1038/ncomms8530
The cell cycle regulator 14-3-3σ opposes and reverses cancer metabolic reprogramming
Abstract
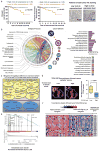
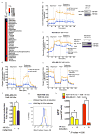
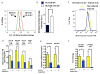
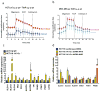
Extensive reprogramming of cellular energy metabolism is a hallmark of cancer. Despite its importance, the molecular mechanism controlling this tumour metabolic shift remains not fully understood. Here we show that 14-3-3σ regulates cancer metabolic reprogramming and protects cells from tumorigenic transformation. 14-3-3σ opposes tumour-promoting metabolic programmes by enhancing c-Myc poly-ubiquitination and subsequent degradation. 14-3-3σ demonstrates the suppressive impact on cancer glycolysis, glutaminolysis, mitochondrial biogenesis and other major metabolic processes of tumours. Importantly, 14-3-3σ expression levels predict overall and recurrence-free survival rates, tumour glucose uptake and metabolic gene expression in breast cancer patients. Thus, these results highlight that 14-3-3σ is an important regulator of tumour metabolism, and loss of 14-3-3σ expression is critical for cancer metabolic reprogramming. We anticipate that pharmacologically elevating the function of 14-3-3σ in tumours could be a promising direction for targeted anticancer metabolism therapy development in future.
Conflict of interest statement
Competing financial interests: The authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

