Differentiation Kinetics of Blood Monocytes and Dendritic Cells in Macaques: Insights to Understanding Human Myeloid Cell Development
- PMID: 26179903
- PMCID: PMC4530075
- DOI: 10.4049/jimmunol.1500522
Differentiation Kinetics of Blood Monocytes and Dendritic Cells in Macaques: Insights to Understanding Human Myeloid Cell Development
Abstract
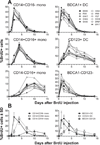
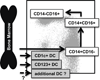
Monocyte and dendritic cell (DC) development was evaluated using in vivo BrdU pulse-chase analyses in rhesus macaques, and phenotype analyses of these cells in blood also were assessed by immunostaining and flow cytometry for comparisons among rhesus, cynomolgus, and pigtail macaques, as well as African green monkeys and humans. The nonhuman primate species and humans have three subsets of monocytes, CD14(+)CD16(-), CD14(+)CD16(+), and CD14(-)CD16(+) cells, which correspond to classical, intermediate, and nonclassical monocytes, respectively. In addition, there exist presently two subsets of DC, BDCA-1(+) myeloid DC and CD123(+) plasmacytoid DC, that were first confirmed in rhesus macaque blood. Following BrdU inoculation, labeled cells first appeared in CD14(+)CD16(-) monocytes, then in CD14(+)CD16(+) cells, and finally in CD14(-)CD16(+) cells, thus defining different stages of monocyte maturation. A fraction of the classical CD14(+)CD16(-) monocytes gradually expressed CD16(+) to become CD16(+)CD14(+) cells and subsequently matured into the nonclassical CD14(-)CD16(+) cell subset. The differentiation kinetics of BDCA-1(+) myeloid DC and CD123(+) plasmacytoid DC were distinct from the monocyte subsets, indicating differences in their myeloid cell origins. Results from studies utilizing nonhuman primates provide valuable information about the turnover, kinetics, and maturation of the different subsets of monocytes and DC using approaches that cannot readily be performed in humans and support further analyses to continue examining the unique myeloid cell origins that may be applied to address disease pathogenesis mechanisms and intervention strategies in humans.
Copyright © 2015 by The American Association of Immunologists, Inc.
Figures

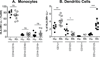



References
-
- Chow A, Brown BD, Merad M. Studying the mononuclear phagocyte system in the molecular age. Nat Rev Immunol. 2011;11:788–798. - PubMed
-
- Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–692. - PubMed
-
- Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, Leenen PJM, Liu Y-J, MacPherson G, Randolph GJ, Scherberich J, Schmitz J, Shortman K, Sozzani S, Strobl H, Zembala M, Austyn JM, Lutz MB. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:e74–e80. - PubMed
-
- Moniuszko M, Bodzenta-Lukaszyk A, Kowal K, Lenczewska D, Dabrowska M. Enhanced frequencies of CD14++CD16+, but not CD14+CD16+, peripheral blood monocytes in severe asthmatic patients. Clin Immunol. 2009;130:338–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 AI110163/AI/NIAID NIH HHS/United States
- R21 AI087302/AI/NIAID NIH HHS/United States
- P51OD011104/OD/NIH HHS/United States
- HL125054/HL/NHLBI NIH HHS/United States
- R01 AI097059/AI/NIAID NIH HHS/United States
- P51 OD011104/OD/NIH HHS/United States
- T32 OD011124/OD/NIH HHS/United States
- R21 AI116198/AI/NIAID NIH HHS/United States
- AI116198/AI/NIAID NIH HHS/United States
- R01 HL125054/HL/NHLBI NIH HHS/United States
- AI110163/AI/NIAID NIH HHS/United States
- AI091501/AI/NIAID NIH HHS/United States
- AI097059/AI/NIAID NIH HHS/United States
- AI087302/AI/NIAID NIH HHS/United States
- R33 AI110163/AI/NIAID NIH HHS/United States
- R21 AI091501/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

