Cutting Edge: A Natural Antisense Transcript, AS-IL1α, Controls Inducible Transcription of the Proinflammatory Cytokine IL-1α
- PMID: 26179904
- PMCID: PMC4530055
- DOI: 10.4049/jimmunol.1500264
Cutting Edge: A Natural Antisense Transcript, AS-IL1α, Controls Inducible Transcription of the Proinflammatory Cytokine IL-1α
Abstract
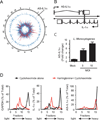
Natural antisense transcripts (NATs) are a class of long noncoding RNAs (lncRNAs) that are complementary to other protein-coding genes. Although thousands of NATs are encoded by mammalian genomes, their functions in innate immunity are unknown. In this study, we identified and characterized a novel NAT, AS-IL1α, which is partially complementary to IL-1α. Similar to IL-1α, AS-IL1α is expressed at low levels in resting macrophages and is induced following infection with Listeria monocytogenes or stimulation with TLR ligands (Pam3CSK4, LPS, polyinosinic-polycytidylic acid). Inducible expression of IL-1α mRNA and protein were significantly reduced in macrophages expressing shRNA that target AS-IL1α. AS-IL1α is located in the nucleus and did not alter the stability of IL-1α mRNA. Instead, AS-IL1α was required for the recruitment of RNA polymerase II to the IL-1α promoter. In summary, our studies identify AS-IL1α as an important regulator of IL-1α transcription during the innate immune response.
Copyright © 2015 by The American Association of Immunologists, Inc.
Figures
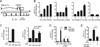

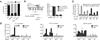
Similar articles
-
Stable Toll-Like Receptor 10 Knockdown in THP-1 Cells Reduces TLR-Ligand-Induced Proinflammatory Cytokine Expression.Int J Mol Sci. 2016 Jun 1;17(6):859. doi: 10.3390/ijms17060859. Int J Mol Sci. 2016. PMID: 27258267 Free PMC article.
-
Knockdown of interleukin-1α does not attenuate LPS-induced production of interleukin-1β in mouse macrophages.Cytokine. 2015 May;73(1):138-43. doi: 10.1016/j.cyto.2015.01.029. Epub 2015 Mar 6. Cytokine. 2015. PMID: 25748836
-
Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis.BMC Mol Biol. 2008 Jan 14;9:6. doi: 10.1186/1471-2199-9-6. BMC Mol Biol. 2008. PMID: 18194570 Free PMC article.
-
Natural antisense and noncoding RNA transcripts as potential drug targets.Drug Discov Today. 2006 Jun;11(11-12):503-8. doi: 10.1016/j.drudis.2006.04.013. Drug Discov Today. 2006. PMID: 16713901 Review.
-
Natural antisense transcript: a concomitant engagement with protein-coding transcript.Oncotarget. 2010 Oct;1(6):447-52. doi: 10.18632/oncotarget.178. Oncotarget. 2010. PMID: 21311100 Free PMC article. Review.
Cited by
-
LncRNAs and immunity: watchdogs for host pathogen interactions.Biol Proced Online. 2017 Apr 27;19:3. doi: 10.1186/s12575-017-0052-7. eCollection 2017. Biol Proced Online. 2017. PMID: 28465674 Free PMC article. Review.
-
Effects of Guchang Capsule on Dextran Sulphate Sodium-Induced Experimental Ulcerative Colitis in Mice.Evid Based Complement Alternat Med. 2016;2016:3150651. doi: 10.1155/2016/3150651. Epub 2016 May 24. Evid Based Complement Alternat Med. 2016. PMID: 27313642 Free PMC article.
-
Long non-coding RNA expressed in macrophage co-varies with the inflammatory phenotype during macrophage development and polarization.J Cell Mol Med. 2019 Oct;23(10):6530-6542. doi: 10.1111/jcmm.14557. Epub 2019 Aug 16. J Cell Mol Med. 2019. PMID: 31419045 Free PMC article. Review.
-
Cytokines and Long Noncoding RNAs.Cold Spring Harb Perspect Biol. 2018 Jun 1;10(6):a028589. doi: 10.1101/cshperspect.a028589. Cold Spring Harb Perspect Biol. 2018. PMID: 28716885 Free PMC article. Review.
-
Biological functions and affected signaling pathways by Long Non-Coding RNAs in the immune system.Noncoding RNA Res. 2024 Sep 6;10:70-90. doi: 10.1016/j.ncrna.2024.09.001. eCollection 2025 Feb. Noncoding RNA Res. 2024. PMID: 39315339 Free PMC article. Review.
References
-
- Consortium, R. G. E. R. G. and G. S. G. (Genome N. P. C. G. and the F. Antisense Transcription in the Mammalian Transcriptome. Science (80-.) 2005;309:1564–1566. - PubMed
-
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

