The development and characterization of synthetic minimal yeast promoters
- PMID: 26183606
- PMCID: PMC4518256
- DOI: 10.1038/ncomms8810
The development and characterization of synthetic minimal yeast promoters
Abstract
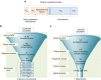
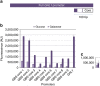
Synthetic promoters, especially minimally sized, are critical for advancing fungal synthetic biology. Fungal promoters often span hundreds of base pairs, nearly ten times the amount of bacterial counterparts. This size limits large-scale synthetic biology efforts in yeasts. Here we address this shortcoming by establishing a methodical workflow necessary to identify robust minimal core elements that can be linked with minimal upstream activating sequences to develop short, yet strong yeast promoters. Through a series of library-based synthesis, analysis and robustness tests, we create a set of non-homologous, purely synthetic, minimal promoters for yeast. These promoters are comprised of short core elements that are generic and interoperable and 10 bp UAS elements that impart strong, constitutive function. Through this methodology, we are able to generate the shortest fungal promoters to date, which can achieve high levels of both inducible and constitutive expression with up to an 80% reduction in size.
Figures



References
-
- Keaveney M. & Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol. Cell 1, 917 (1998). - PubMed
-
- Teo W. S. & Chang M. W. Development and characterization of AND-gate dynamic controllers with a modular synthetic GAL1 core promoter in Saccharomyces cerevisiae. Biotechnol. Bioeng. 111, 144 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

