Control of mRNA decapping by positive and negative regulatory elements in the Dcp2 C-terminal domain
- PMID: 26184073
- PMCID: PMC4536323
- DOI: 10.1261/rna.052449.115
Control of mRNA decapping by positive and negative regulatory elements in the Dcp2 C-terminal domain
Abstract
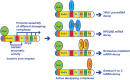
Decapping commits an mRNA to complete degradation and promotes general 5' to 3' decay, nonsense-mediated decay (NMD), and transcript-specific degradation. In Saccharomyces cerevisiae, a single decapping enzyme composed of a regulatory subunit (Dcp1) and a catalytic subunit (Dcp2) targets thousands of distinct substrate mRNAs. However, the mechanisms controlling this enzyme's in vivo activity and substrate specificity remain elusive. Here, using a genetic approach, we show that the large C-terminal domain of Dcp2 includes a set of conserved negative and positive regulatory elements. A single negative element inhibits enzymatic activity and controls the downstream functions of several positive elements. The positive elements recruit the specific decapping activators Edc3, Pat1, and Upf1 to form distinct decapping complexes and control the enzyme's substrate specificity and final activation. Our results reveal unforeseen regulatory mechanisms that control decapping enzyme activity and function in vivo, and define roles for several decapping activators in the regulation of mRNA decapping.
Keywords: decapping activators; decapping enzyme; mRNA decapping; positive and negative regulation.
© 2015 He and Jacobson; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





References
-
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A. 2004. Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell 15: 5–15. - PubMed
-
- Bashor CJ, Helman NC, Yan S, Lim WA. 2008. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science 319: 1539–1543. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
