An integrative approach to analyze microarray datasets for prioritization of genes relevant to lens biology and disease
- PMID: 26185746
- PMCID: PMC4500531
- DOI: 10.1016/j.gdata.2015.06.017
An integrative approach to analyze microarray datasets for prioritization of genes relevant to lens biology and disease
Abstract
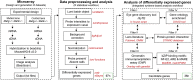
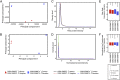
Microarray-based profiling represents an effective method to analyze cellular or tissue-specific gene expression on the genome-level. However, in comparative analyses between control and mutant samples, microarrays often identify a large number of differentially expressed genes, in turn making it challenging to isolate the select "high-priority candidates" that are most relevant to an observed mutant phenotype. Here, we describe an integrative approach for mouse mutant lens microarray gene expression analysis using publically accessible systems-level information such as wild-type mouse lens expression data in iSyTE (integrated Systems Tool for Eye gene discovery), protein-protein interaction data in public databases, gene ontology enrichment data, and transcription factor binding profile data. This strategy, when applied to small Maf Mafg-/-:Mafk+/- mouse lens microarray datasets (deposited in NCBI Gene Expression Omnibus database with accession number GSE65500) in Agrawal et al. 2015 [1], led to the effective prioritization of candidate genes linked to lens defects in these mutants. Indeed, from the original list of genes that are differentially expressed at ±1.5-fold and p<0.05 in Mafg-/-:Mafk+/- mutant lenses, this analysis led to the identification of thirty-six high-priority candidates, in turn reducing the number of genes for further study by approximately 1/3rd of the total. Moreover, eight of these genes are linked to mammalian cataract in the published literature, validating the efficacy of this approach. Additionally, these high-priority candidates contribute valuable information for the assembly of a gene regulatory network in the lens. In sum, the pipeline outlined in this report represents an effective approach for initial as well as downstream microarray expression data analysis to identify genes important for lens biology and cataracts. We anticipate that this integrative strategy can be extended to prioritize phenotypically relevant candidate genes from microarray data in other cells and tissues.
Keywords: Cataract; Gene expression; Lens; Microarrays; iSyTE.
Figures

References
-
- Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. - PubMed
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300.
-
- Huang D.W., Sherman B.T., Zheng X., Yang J., Imamichi T., Stephens R. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinformatics. 2009 (Unit 13.11. Chapter 13) - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
