Elucidating Compound Mechanism of Action by Network Perturbation Analysis
- PMID: 26186195
- PMCID: PMC4506491
- DOI: 10.1016/j.cell.2015.05.056
Elucidating Compound Mechanism of Action by Network Perturbation Analysis
Abstract
Genome-wide identification of the mechanism of action (MoA) of small-molecule compounds characterizing their targets, effectors, and activity modulators represents a highly relevant yet elusive goal, with critical implications for assessment of compound efficacy and toxicity. Current approaches are labor intensive and mostly limited to elucidating high-affinity binding target proteins. We introduce a regulatory network-based approach that elucidates genome-wide MoA proteins based on the assessment of the global dysregulation of their molecular interactions following compound perturbation. Analysis of cellular perturbation profiles identified established MoA proteins for 70% of the tested compounds and elucidated novel proteins that were experimentally validated. Finally, unknown-MoA compound analysis revealed altretamine, an anticancer drug, as an inhibitor of glutathione peroxidase 4 lipid repair activity, which was experimentally confirmed, thus revealing unexpected similarity to the activity of sulfasalazine. This suggests that regulatory network analysis can provide valuable mechanistic insight into the elucidation of small-molecule MoA and compound similarity.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

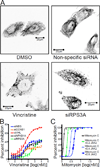
Comment in
-
Systems biology: MoA on DeMAND.Nat Rev Genet. 2015 Sep;16(9):498-9. doi: 10.1038/nrg3996. Epub 2015 Aug 4. Nat Rev Genet. 2015. PMID: 26239713 No abstract available.
References
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
-
- Bansal M, Della Gatta G, di Bernardo D. Inference of gene regulatory networks and compound mode of action from time course gene expression profiles. Bioinformatics. 2006;22:815–822. - PubMed
-
- Basilico C, Pennacchietti S, Vigna E, Chiriaco C, Arena S, Bardelli A, Valdembri D, Serini G, Michieli P. Tivantinib (ARQ197) Displays Cytotoxic Activity That Is Independent of Its Ability to Bind MET. Clinical Cancer Research. 2013;19:2381–2392. - PubMed
-
- Basso K, Saito M, Sumazin P, Margolin AA, Wang K, Lim WK, Kitagawa Y, Schneider C, Alvarez MJ, Califano A, et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115:975–984. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 5U54CA121852-08/CA/NCI NIH HHS/United States
- R35 CA209896/CA/NCI NIH HHS/United States
- U01 CA168426/CA/NCI NIH HHS/United States
- 5R01CA097061/CA/NCI NIH HHS/United States
- U01 HL111566/HL/NHLBI NIH HHS/United States
- R01 CA161061/CA/NCI NIH HHS/United States
- S10 OD012351/OD/NIH HHS/United States
- U54 CA121852/CA/NCI NIH HHS/United States
- R01 CA097061/CA/NCI NIH HHS/United States
- 3U01HL111566-02/HL/NHLBI NIH HHS/United States
- U01 CA164184/CA/NCI NIH HHS/United States
- R01CA161061/CA/NCI NIH HHS/United States
- Howard Hughes Medical Institute/United States
- 5U01CA168426/CA/NCI NIH HHS/United States
- 1U01CA164184-02/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

