Tempo and Mode of Transposable Element Activity in Drosophila
- PMID: 26186437
- PMCID: PMC4505896
- DOI: 10.1371/journal.pgen.1005406
Tempo and Mode of Transposable Element Activity in Drosophila
Abstract
The evolutionary dynamics of transposable element (TE) insertions have been of continued interest since TE activity has important implications for genome evolution and adaptation. Here, we infer the transposition dynamics of TEs by comparing their abundance in natural D. melanogaster and D. simulans populations. Sequencing pools of more than 550 South African flies to at least 320-fold coverage, we determined the genome wide TE insertion frequencies in both species. We suggest that the predominance of low frequency insertions in the two species (>80% of the insertions have a frequency <0.2) is probably due to a high activity of more than 58 families in both species. We provide evidence for 50% of the TE families having temporally heterogenous transposition rates with different TE families being affected in the two species. While in D. melanogaster retrotransposons were more active, DNA transposons showed higher activity levels in D. simulans. Moreover, we suggest that LTR insertions are mostly of recent origin in both species, while DNA and non-LTR insertions are older and more frequently vertically transmitted since the split of D. melanogaster and D. simulans. We propose that the high TE activity is of recent origin in both species and a consequence of the demographic history, with habitat expansion triggering a period of rapid evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


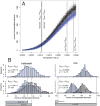
References
-
- Charlesworth B, Charlesworth D. The population dynamics of transposable elements. Genetical Research. 1983;42(01):1–27.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

