A Conserved Histidine in the RNA Sensor RIG-I Controls Immune Tolerance to N1-2'O-Methylated Self RNA
- PMID: 26187414
- PMCID: PMC7128463
- DOI: 10.1016/j.immuni.2015.06.015
A Conserved Histidine in the RNA Sensor RIG-I Controls Immune Tolerance to N1-2'O-Methylated Self RNA
Abstract
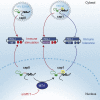
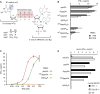
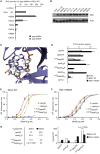
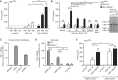
The cytosolic helicase retinoic acid-inducible gene-I (RIG-I) initiates immune responses to most RNA viruses by detecting viral 5'-triphosphorylated RNA (pppRNA). Although endogenous mRNA is also 5'-triphosphorylated, backbone modifications and the 5'-ppp-linked methylguanosine ((m7)G) cap prevent immunorecognition. Here we show that the methylation status of endogenous capped mRNA at the 5'-terminal nucleotide (N1) was crucial to prevent RIG-I activation. Moreover, we identified a single conserved amino acid (H830) in the RIG-I RNA binding pocket as the mediator of steric exclusion of N1-2'O-methylated RNA. H830A alteration (RIG-I(H830A)) restored binding of N1-2'O-methylated pppRNA. Consequently, endogenous mRNA activated the RIG-I(H830A) mutant but not wild-type RIG-I. Similarly, knockdown of the endogenous N1-2'O-methyltransferase led to considerable RIG-I stimulation in the absence of exogenous stimuli. Studies involving yellow-fever-virus-encoded 2'O-methyltransferase and RIG-I(H830A) revealed that viruses exploit this mechanism to escape RIG-I. Our data reveal a new role for cap N1-2'O-methylation in RIG-I tolerance of self-RNA.
Keywords: 2′O-methyl; 5′-triphosphate RNA; MTr1; RIG-I; cap; immune recognition of RNA; innate immune tolerance mechanism; mRNA; virus.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
Comment in
-
A Sense of Self: RIG-I's Tolerance to Host RNA.Immunity. 2015 Jul 21;43(1):1-2. doi: 10.1016/j.immuni.2015.06.022. Immunity. 2015. PMID: 26200004
References
-
- Bray M. Highly pathogenic RNA viral infections: challenges for antiviral research. Antiviral Res. 2008;78:1–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

