Regulating the Regulators: Recent Revelations in the Control of E3 Ubiquitin Ligases
- PMID: 26187467
- PMCID: PMC4571856
- DOI: 10.1074/jbc.R115.675165
Regulating the Regulators: Recent Revelations in the Control of E3 Ubiquitin Ligases
Abstract
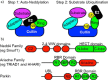
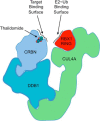
Since its discovery as a post-translational signal for protein degradation, our understanding of ubiquitin (Ub) has vastly evolved. Today, we recognize that the role of Ub signaling is expansive and encompasses diverse processes including cell division, the DNA damage response, cellular immune signaling, and even organismal development. With such a wide range of functions comes a wide range of regulatory mechanisms that control the activity of the ubiquitylation machinery. Ub attachment to substrates occurs through the sequential action of three classes of enzymes, E1s, E2s, and E3s. In humans, there are 2 E1s, ∼ 35 E2s, and hundreds of E3s that work to attach Ub to thousands of cellular substrates. Regulation of ubiquitylation can occur at each stage of the stepwise Ub transfer process, and substrates can also impact their own modification. Recent studies have revealed elegant mechanisms that have evolved to control the activity of the enzymes involved. In this minireview, we highlight recent discoveries that define some of the various mechanisms by which the activities of E3-Ub ligases are regulated.
Keywords: E3 ubiquitin ligase; HECT; RING; cullin; parkin; ubiquitin; ubiquitin ligase; ubiquitin regulation; ubiquitylation (ubiquitination).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

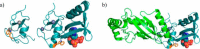
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

