Genome sequence and comparative analysis of a putative entomopathogenic Serratia isolated from Caenorhabditis briggsae
- PMID: 26187596
- PMCID: PMC4506600
- DOI: 10.1186/s12864-015-1697-8
Genome sequence and comparative analysis of a putative entomopathogenic Serratia isolated from Caenorhabditis briggsae
Abstract
Background: Entomopathogenic associations between nematodes in the genera Steinernema and Heterorhabdus with their cognate bacteria from the bacterial genera Xenorhabdus and Photorhabdus, respectively, are extensively studied for their potential as biological control agents against invasive insect species. These two highly coevolved associations were results of convergent evolution. Given the natural abundance of bacteria, nematodes and insects, it is surprising that only these two associations with no intermediate forms are widely studied in the entomopathogenic context. Discovering analogous systems involving novel bacterial and nematode species would shed light on the evolutionary processes involved in the transition from free living organisms to obligatory partners in entomopathogenicity.
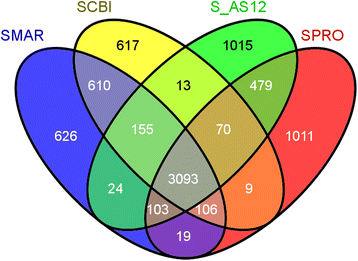
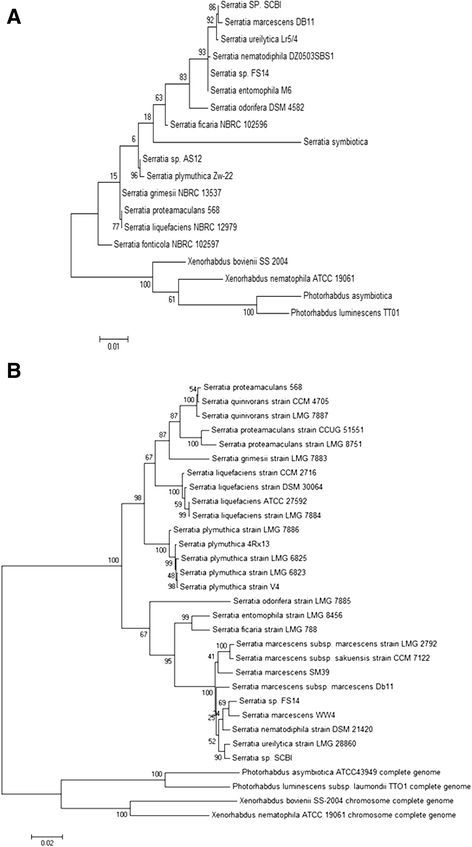
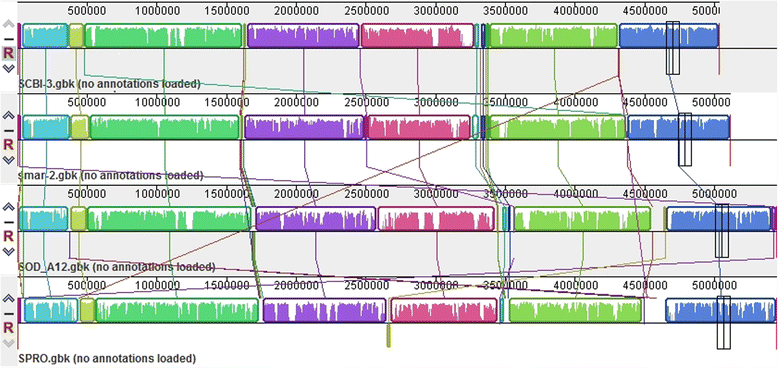
Results: We report the complete genome sequence of a new member of the enterobacterial genus Serratia that forms a putative entomopathogenic complex with Caenorhabditis briggsae. Analysis of the 5.04 MB chromosomal genome predicts 4599 protein coding genes, seven sets of ribosomal RNA genes, 84 tRNA genes and a 64.8 KB plasmid encoding 74 genes. Comparative genomic analysis with three of the previously sequenced Serratia species, S. marcescens DB11 and S. proteamaculans 568, and Serratia sp. AS12, revealed that these four representatives of the genus share a core set of ~3100 genes and extensive structural conservation. The newly identified species shares a more recent common ancestor with S. marcescens with 99% sequence identity in rDNA sequence and orthology across 85.6% of predicted genes. Of the 39 genes/operons implicated in the virulence, symbiosis, recolonization, immune evasion and bioconversion, 21 (53.8%) were present in Serratia while 33 (84.6%) and 35 (89%) were present in Xenorhabdus and Photorhabdus EPN bacteria respectively.
Conclusion: The majority of unique sequences in Serratia sp. SCBI (South African Caenorhabditis briggsae Isolate) are found in ~29 genomic islands of 5 to 65 genes and are enriched in putative functions that are biologically relevant to an entomopathogenic lifestyle, including non-ribosomal peptide synthetases, bacteriocins, fimbrial biogenesis, ushering proteins, toxins, secondary metabolite secretion and multiple drug resistance/efflux systems. By revealing the early stages of adaptation to this lifestyle, the Serratia sp. SCBI genome underscores the fact that in EPN formation the composite end result - killing, bioconversion, cadaver protection and recolonization- can be achieved by dissimilar mechanisms. This genome sequence will enable further study of the evolution of entomopathogenic nematode-bacteria complexes.
Figures


References
-
- Allen AE, CL Dupont, M Obornik, A Horak, A Nunes-Nesi, JP McCrow, et al. Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature. 2011;473(7346):203−+. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

