FANCD2 and REV1 cooperate in the protection of nascent DNA strands in response to replication stress
- PMID: 26187992
- PMCID: PMC4787816
- DOI: 10.1093/nar/gkv737
FANCD2 and REV1 cooperate in the protection of nascent DNA strands in response to replication stress
Abstract
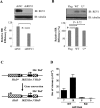
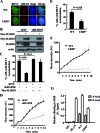
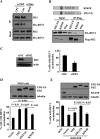
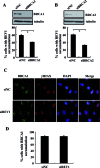
REV1 is a eukaryotic member of the Y-family of DNA polymerases involved in translesion DNA synthesis and genome mutagenesis. Recently, REV1 is also found to function in homologous recombination. However, it remains unclear how REV1 is recruited to the sites where homologous recombination is processed. Here, we report that loss of mammalian REV1 results in a specific defect in replication-associated gene conversion. We found that REV1 is targeted to laser-induced DNA damage stripes in a manner dependent on its ubiquitin-binding motifs, on RAD18, and on monoubiquitinated FANCD2 (FANCD2-mUb) that associates with REV1. Expression of a FANCD2-Ub chimeric protein in RAD18-depleted cells enhances REV1 assembly at laser-damaged sites, suggesting that FANCD2-mUb functions downstream of RAD18 to recruit REV1 to DNA breaks. Consistent with this suggestion we found that REV1 and FANCD2 are epistatic with respect to sensitivity to the double-strand break-inducer camptothecin. REV1 enrichment at DNA damage stripes also partially depends on BRCA1 and BRCA2, components of the FANCD2/BRCA supercomplex. Intriguingly, analogous to FANCD2-mUb and BRCA1/BRCA2, REV1 plays an unexpected role in protecting nascent replication tracts from degradation by stabilizing RAD51 filaments. Collectively these data suggest that REV1 plays multiple roles at stalled replication forks in response to replication stress.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
FANC pathway promotes UV-induced stalled replication forks recovery by acting both upstream and downstream Polη and Rev1.PLoS One. 2013;8(1):e53693. doi: 10.1371/journal.pone.0053693. Epub 2013 Jan 24. PLoS One. 2013. PMID: 23365640 Free PMC article.
-
CtIP mediates replication fork recovery in a FANCD2-regulated manner.Hum Mol Genet. 2014 Jul 15;23(14):3695-705. doi: 10.1093/hmg/ddu078. Epub 2014 Feb 20. Hum Mol Genet. 2014. PMID: 24556218 Free PMC article.
-
Fanconi anemia complementation group D2 (FANCD2) functions independently of BRCA2- and RAD51-associated homologous recombination in response to DNA damage.J Biol Chem. 2005 Apr 15;280(15):14877-83. doi: 10.1074/jbc.M414669200. Epub 2005 Jan 25. J Biol Chem. 2005. PMID: 15671039
-
Beyond interstrand crosslinks repair: contribution of FANCD2 and other Fanconi Anemia proteins to the replication of DNA.Mutat Res. 2018 Mar;808:83-92. doi: 10.1016/j.mrfmmm.2017.09.004. Epub 2017 Sep 14. Mutat Res. 2018. PMID: 29031493 Review.
-
How Fanconi anemia proteins promote the four Rs: replication, recombination, repair, and recovery.Environ Mol Mutagen. 2005 Mar-Apr;45(2-3):128-42. doi: 10.1002/em.20109. Environ Mol Mutagen. 2005. PMID: 15668941 Review.
Cited by
-
REV1 promotes lung tumorigenesis by activating the Rad18/SERTAD2 axis.Cell Death Dis. 2022 Feb 3;13(2):110. doi: 10.1038/s41419-022-04567-5. Cell Death Dis. 2022. PMID: 35115490 Free PMC article.
-
WRNIP1 Protects Reversed DNA Replication Forks from SLX4-Dependent Nucleolytic Cleavage.iScience. 2019 Nov 22;21:31-41. doi: 10.1016/j.isci.2019.10.010. Epub 2019 Oct 8. iScience. 2019. PMID: 31654852 Free PMC article.
-
Drug Discovery Targeting Post-Translational Modifications in Response to DNA Damages Induced by Space Radiation.Int J Mol Sci. 2023 Apr 21;24(8):7656. doi: 10.3390/ijms24087656. Int J Mol Sci. 2023. PMID: 37108815 Free PMC article. Review.
-
Polη O-GlcNAcylation governs genome integrity during translesion DNA synthesis.Nat Commun. 2017 Dec 5;8(1):1941. doi: 10.1038/s41467-017-02164-1. Nat Commun. 2017. PMID: 29208956 Free PMC article.
-
Regulation of DNA repair by non-coding miRNAs.Noncoding RNA Res. 2016 Nov 5;1(1):64-68. doi: 10.1016/j.ncrna.2016.10.002. eCollection 2016 Oct. Noncoding RNA Res. 2016. PMID: 30159412 Free PMC article. Review.
References
-
- Tissier A., Kannouche P., Reck M.P., Lehmann A.R., Fuchs R.P., Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair. 2004;3:1503–1514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

