Precision metabolic engineering: The design of responsive, selective, and controllable metabolic systems
- PMID: 26189665
- PMCID: PMC4770000
- DOI: 10.1016/j.ymben.2015.06.011
Precision metabolic engineering: The design of responsive, selective, and controllable metabolic systems
Abstract
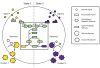
Metabolic engineering is generally focused on static optimization of cells to maximize production of a desired product, though recently dynamic metabolic engineering has explored how metabolic programs can be varied over time to improve titer. However, these are not the only types of applications where metabolic engineering could make a significant impact. Here, we discuss a new conceptual framework, termed "precision metabolic engineering," involving the design and engineering of systems that make different products in response to different signals. Rather than focusing on maximizing titer, these types of applications typically have three hallmarks: sensing signals that determine the desired metabolic target, completely directing metabolic flux in response to those signals, and producing sharp responses at specific signal thresholds. In this review, we will first discuss and provide examples of precision metabolic engineering. We will then discuss each of these hallmarks and identify which existing metabolic engineering methods can be applied to accomplish those tasks, as well as some of their shortcomings. Ultimately, precise control of metabolic systems has the potential to enable a host of new metabolic engineering and synthetic biology applications for any problem where flexibility of response to an external signal could be useful.
Keywords: Metabolic control; Pathway regulation; Precision metabolic engineering; Product selectivity; Sensory systems; Synthetic biology.
Copyright © 2015 International Metabolic Engineering Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Alonso-Gutierrez J, Chan R, Batth TS, Adams PD, Keasling JD, Petzold CJ, Lee TS. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab Eng. 2013;19:33–41. - PubMed
-
- Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol. 2005a;23:612–616. - PubMed
-
- Anesiadis N, Cluett WR, Mahadevan R. Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng. 2008;10:255–266. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

