RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4
- PMID: 26189797
- PMCID: PMC4820683
- DOI: 10.1038/onc.2015.235
RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4
Abstract
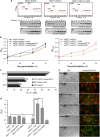
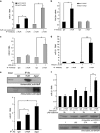
Translation control of proinflammatory genes has a crucial role in regulating the inflammatory response and preventing chronic inflammation, including a transition to cancer. The proinflammatory tumor suppressor protein programmed cell death 4 (PDCD4) is important for maintaining the balance between inflammation and tumorigenesis. PDCD4 messenger RNA translation is inhibited by the oncogenic microRNA, miR-21. AU-rich element-binding protein HuR was found to interact with the PDCD4 3'-untranslated region (UTR) and prevent miR-21-mediated repression of PDCD4 translation. Cells stably expressing miR-21 showed higher proliferation and reduced apoptosis, which was reversed by HuR expression. Inflammatory stimulus caused nuclear-cytoplasmic relocalization of HuR, reversing the translation repression of PDCD4. Unprecedentedly, HuR was also found to bind to miR-21 directly, preventing its interaction with the PDCD4 3'-UTR, thereby preventing the translation repression of PDCD4. This suggests that HuR might act as a 'miRNA sponge' to regulate miRNA-mediated translation regulation under conditions of stress-induced nuclear-cytoplasmic translocation of HuR, which would allow fine-tuned gene expression in complex regulatory environments.
Figures





References
-
- Lankat-Buttgereit B, Göke R. The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol Cell 2009; 101: 309–317. - PubMed
-
- Hilliard A, Hilliard B, Zheng S-J, Sun H, Miwa T, Song W et al. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J Immunol 2006; 177: 8095–8102. - PubMed
-
- Biyanee A, Ohnheiser J, Singh P, Klempnauer K-H. A novel mechanism for the control of translation of specific mRNAs by tumor suppressor protein Pdcd4: inhibition of translation elongation. Oncogene 2014; 34: 1384–1392. - PubMed
-
- Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol 2010; 10: 24–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

