Targeted Regression of Hepatocellular Carcinoma by Cancer-Specific RNA Replacement through MicroRNA Regulation
- PMID: 26189916
- PMCID: PMC4507181
- DOI: 10.1038/srep12315
Targeted Regression of Hepatocellular Carcinoma by Cancer-Specific RNA Replacement through MicroRNA Regulation
Abstract
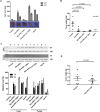
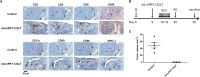
Hepatocellular carcinoma (HCC) has a high fatality rate and limited therapeutic options with side effects and low efficacy. Here, we proposed a new anti-HCC approach based on cancer-specific post-transcriptional targeting. To this end, trans-splicing ribozymes from Tetrahymena group I intron were developed, which can specifically induce therapeutic gene activity through HCC-specific replacement of telomerase reverse transcriptase (TERT) RNA. To circumvent side effects due to TERT expression in regenerating liver tissue, liver-specific microRNA-regulated ribozymes were constructed by incorporating complementary binding sites for the hepatocyte-selective microRNA-122a (miR-122a), which is down-regulated in HCC. The ribozyme activity in vivo was assessed in mouse models orthotopically implanted with HCC. Systemic administration of adenovirus encoding the developed ribozymes caused efficient anti-cancer effect and the least hepatotoxicity with regulation of ribozyme expression by miR-122a in both xenografted and syngeneic orthotopic murine model of multifocal HCC. Of note, the ribozyme induced local and systemic antitumor immunity, thereby completely suppressing secondary tumor challenge in the syngeneic mouse. The cancer specific trans-splicing ribozyme system, which mediates tissue-specific microRNA-regulated RNA replacement, provides a clinically relevant, safe, and efficient strategy for HCC treatment.
Figures




References
-
- Monsour H. P. Jr et al. Hepatocellular carcinoma, the rising tide from east to west-are view of epidemiology, screening and tumor markers. Transl. Cancer Res. 2, 492–506 (2013).
-
- McCormick F. Cancer gene therapy, fringe or cutting edge. Nat. Rev. Cancer. 1, 130–141 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

