Widespread Inducible Transcription Downstream of Human Genes
- PMID: 26190259
- PMCID: PMC4530028
- DOI: 10.1016/j.molcel.2015.06.016
Widespread Inducible Transcription Downstream of Human Genes
Abstract
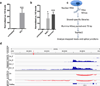
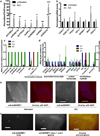
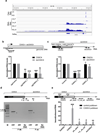
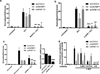
Pervasive transcription of the human genome generates RNAs whose mode of formation and functions are largely uncharacterized. Here, we combine RNA-seq with detailed mechanistic studies to describe a transcript type derived from protein-coding genes. The resulting RNAs, which we call DoGs for downstream of gene containing transcripts, possess long non-coding regions (often >45 kb) and remain chromatin bound. DoGs are inducible by osmotic stress through an IP3 receptor signaling-dependent pathway, indicating active regulation. DoG levels are increased by decreased termination of the upstream transcript, a previously undescribed mechanism for rapid transcript induction. Relative depletion of polyA signals in DoG regions correlates with increased levels of DoGs after osmotic stress. We detect DoG transcription in several human cell lines and provide evidence for thousands of DoGs genome wide.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Transcription: Stress unleashes the DoGs.Nat Rev Genet. 2015 Sep;16(9):497. doi: 10.1038/nrg3994. Epub 2015 Aug 4. Nat Rev Genet. 2015. PMID: 26239712 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

