The spatial biology of transcription and translation in rapidly growing Escherichia coli
- PMID: 26191045
- PMCID: PMC4488752
- DOI: 10.3389/fmicb.2015.00636
The spatial biology of transcription and translation in rapidly growing Escherichia coli
Abstract
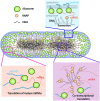
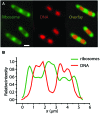
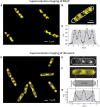
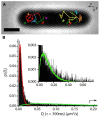
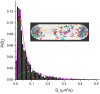
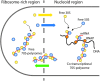
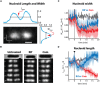
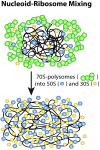
Single-molecule fluorescence provides high resolution spatial distributions of ribosomes and RNA polymerase (RNAP) in live, rapidly growing Escherichia coli. Ribosomes are more strongly segregated from the nucleoids (chromosomal DNA) than previous widefield fluorescence studies suggested. While most transcription may be co-translational, the evidence indicates that most translation occurs on free mRNA copies that have diffused from the nucleoids to a ribosome-rich region. Analysis of time-resolved images of the nucleoid spatial distribution after treatment with the transcription-halting drug rifampicin and the translation-halting drug chloramphenicol shows that both drugs cause nucleoid contraction on the 0-3 min timescale. This is consistent with the transertion hypothesis. We suggest that the longer-term (20-30 min) nucleoid expansion after Rif treatment arises from conversion of 70S-polysomes to 30S and 50S subunits, which readily penetrate the nucleoids. Monte Carlo simulations of a polymer bead model built to mimic the chromosomal DNA and ribosomes (either 70S-polysomes or 30S and 50S subunits) explain spatial segregation or mixing of ribosomes and nucleoids in terms of excluded volume and entropic effects alone. A comprehensive model of the transcription-translation-transertion system incorporates this new information about the spatial organization of the E. coli cytoplasm. We propose that transertion, which radially expands the nucleoids, is essential for recycling of 30S and 50S subunits from ribosome-rich regions back into the nucleoids. There they initiate co-transcriptional translation, which is an important mechanism for maintaining RNAP forward progress and protecting the nascent mRNA chain. Segregation of 70S-polysomes from the nucleoid may facilitate rapid growth by shortening the search time for ribosomes to find free mRNA concentrated outside the nucleoid and the search time for RNAP concentrated within the nucleoid to find transcription initiation sites.
Keywords: DNA-ribosome spatial segregation; E. coli; RNA polymerase; nucleoid structure; ribosomes; single-molecule tracking live cell.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

