Phenolyzer: phenotype-based prioritization of candidate genes for human diseases
- PMID: 26192085
- PMCID: PMC4718403
- DOI: 10.1038/nmeth.3484
Phenolyzer: phenotype-based prioritization of candidate genes for human diseases
Abstract
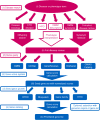
Prior biological knowledge and phenotype information may help to identify disease genes from human whole-genome and whole-exome sequencing studies. We developed Phenolyzer (http://phenolyzer.usc.edu), a tool that uses prior information to implicate genes involved in diseases. Phenolyzer exhibits superior performance over competing methods for prioritizing Mendelian and complex disease genes, based on disease or phenotype terms entered as free text.
Conflict of interest statement
The authors declare competing financial interests: details are available in the online version of the paper.
Figures
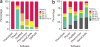
Similar articles
-
An automatic and efficient pipeline for disease gene identification through utilizing family-based sequencing data.Biomed Mater Eng. 2015;26 Suppl 1:S1797-803. doi: 10.3233/BME-151481. Biomed Mater Eng. 2015. PMID: 26405949
-
[Diagnostics in human genetics : Integration of phenotypic and genomic data].Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2017 May;60(5):542-549. doi: 10.1007/s00103-017-2538-5. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2017. PMID: 28293716 German.
-
Phen-Gen: combining phenotype and genotype to analyze rare disorders.Nat Methods. 2014 Sep;11(9):935-7. doi: 10.1038/nmeth.3046. Epub 2014 Aug 3. Nat Methods. 2014. PMID: 25086502
-
Exome sequencing greatly expedites the progressive research of Mendelian diseases.Front Med. 2014 Mar;8(1):42-57. doi: 10.1007/s11684-014-0303-9. Epub 2014 Jan 3. Front Med. 2014. PMID: 24384736 Review.
-
Computational and bioinformatics frameworks for next-generation whole exome and genome sequencing.ScientificWorldJournal. 2013;2013:730210. doi: 10.1155/2013/730210. Epub 2013 Jan 13. ScientificWorldJournal. 2013. PMID: 23365548 Free PMC article. Review.
Cited by
-
Detection of a novel SETBP1 variant in a Chinese neonate with Schinzel-Giedion syndrome.Front Pediatr. 2022 Sep 6;10:920741. doi: 10.3389/fped.2022.920741. eCollection 2022. Front Pediatr. 2022. PMID: 36147799 Free PMC article.
-
Novel lissencephaly-associated NDEL1 variant reveals distinct roles of NDE1 and NDEL1 in nucleokinesis and human cortical malformations.Acta Neuropathol. 2024 Jan 9;147(1):13. doi: 10.1007/s00401-023-02665-y. Acta Neuropathol. 2024. PMID: 38194050 Free PMC article.
-
Phenotype-genotype comorbidity analysis of patients with rare disorders provides insight into their pathological and molecular bases.PLoS Genet. 2020 Oct 1;16(10):e1009054. doi: 10.1371/journal.pgen.1009054. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33001999 Free PMC article.
-
Genetic characteristics of common variable immunodeficiency patients with autoimmunity.Front Genet. 2023 Nov 13;14:1209988. doi: 10.3389/fgene.2023.1209988. eCollection 2023. Front Genet. 2023. PMID: 38028622 Free PMC article.
-
An Analysis of the Pathogenic Genes and Mutation Sites of Macrodactyly.Pharmgenomics Pers Med. 2022 Jan 29;15:55-64. doi: 10.2147/PGPM.S346373. eCollection 2022. Pharmgenomics Pers Med. 2022. PMID: 35125881 Free PMC article.
References
-
- Jäger M, et al. Hum Mutat. 2014;35:548–555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

